A Deep Dive into Bioinformatics Tools and Databases for Genetic Discovery
November 16, 2023I. Introduction to Bioinformatics
A. Definition and Scope
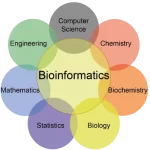
Bioinformatics is an interdisciplinary field that combines biology and computer science to analyze and interpret biological data. It involves the application of computational and statistical techniques to the understanding and management of biological information. The scope of bioinformatics extends to various aspects of biological research, including genomics, proteomics, structural biology, evolutionary biology, and more.
Bioinformatics encompasses the development and application of tools and algorithms for storing, retrieving, organizing, and analyzing biological data. It plays a crucial role in the era of big data in biology, where the amount of biological information generated through technologies like DNA sequencing and high-throughput experimentation is vast and complex.
B. Importance in Biological Research
- Data Management: Bioinformatics facilitates the storage and retrieval of massive biological datasets. This includes DNA sequences, protein structures, gene expression profiles, and more. Efficient data management is essential for organizing and accessing this wealth of information.
- Genome Sequencing and Annotation: With the advent of high-throughput sequencing technologies, bioinformatics has become indispensable in the analysis and interpretation of genomic data. It aids in the identification and annotation of genes, regulatory elements, and other functional elements within genomes.
- Comparative Genomics: Bioinformatics enables the comparison of genetic information across different species. Comparative genomics helps identify evolutionarily conserved elements, understand genetic variation, and gain insights into the functional significance of genes.
- Proteomics: Bioinformatics plays a key role in the analysis of protein structures and functions. It aids in the identification of protein-protein interactions, prediction of protein structures, and annotation of protein functions.
- Structural Biology: Bioinformatics tools are used to analyze and predict the three-dimensional structures of biological macromolecules. This is crucial for understanding the relationship between structure and function in proteins, nucleic acids, and other biomolecules.
C. Role in Genetic Discovery and Analysis
- Functional Genomics: Bioinformatics tools contribute to the understanding of gene function on a genome-wide scale. This involves the analysis of gene expression, regulation, and the functional consequences of genetic variation.
- Pharmacogenomics: Bioinformatics is applied to study the relationship between an individual’s genetic makeup and their response to drugs. This helps in the development of personalized medicine, tailoring treatments based on the patient’s genetic profile.
- Disease Biomarker Discovery: Bioinformatics is employed in the identification of molecular markers associated with diseases. This has implications for early diagnosis, prognosis, and the development of targeted therapies.
In summary, bioinformatics is a critical field that bridges biology and computational science, providing essential tools for the analysis and interpretation of biological data. Its applications in genomics, proteomics, and genetic analysis have significantly contributed to advancements in biological research and have practical implications in medicine and biotechnology.
II. Essential Bioinformatics Tools
A. Sequence Analysis Tools
- BLAST (Basic Local Alignment Search Tool):
- Function: BLAST is a widely used tool for comparing biological sequences, such as DNA, RNA, or protein sequences, against a database to identify homologous sequences.
- Application: It is crucial for tasks like sequence similarity searching, functional annotation of genes, and identifying evolutionary relationships between sequences.
- Features: BLAST provides different algorithms (e.g., BLASTp for protein sequences, BLASTn for nucleotide sequences) and allows users to customize search parameters to balance sensitivity and specificity.
- Clustal Omega for Multiple Sequence Alignment:
- Function: Clustal Omega is a tool for multiple sequence alignment, where it aligns three or more biological sequences to identify regions of similarity.
- Application: Multiple sequence alignment is essential for understanding the evolutionary relationships between sequences, identifying conserved regions, and predicting functional domains in proteins.
- Features: Clustal Omega is known for its speed and scalability, making it suitable for aligning large datasets. It provides options for visualization and analysis of the alignment results.
- HMMER for Protein Sequence Analysis:
- Function: HMMER (Hidden Markov Model based on Evolutionary Relationships) is used for searching sequence databases for homologous protein sequences, incorporating the statistical framework of hidden Markov models.
- Application: HMMER is particularly useful for identifying remote homologs and annotating protein families or domains.
- Features: It allows the construction of custom hidden Markov models, providing a more sensitive approach to detect relationships between protein sequences. HMMER is employed in the annotation of functional domains and identification of conserved motifs.
These sequence analysis tools are fundamental in bioinformatics, providing researchers with the means to compare, align, and analyze biological sequences. They play a pivotal role in tasks such as functional annotation, evolutionary analysis, and understanding the structure-function relationships of biological macromolecules. Researchers often integrate these tools into their workflows to gain insights into the vast amount of biological sequence data generated by modern experimental techniques.
B. Structural Analysis Tools
- PyMOL for Molecular Visualization:
- Function: PyMOL is a powerful molecular visualization tool that allows users to create high-quality 3D images and animations of molecular structures.
- Application: Researchers use PyMOL to visualize and analyze macromolecular structures, such as proteins, nucleic acids, and small molecules. It aids in understanding the spatial arrangement of atoms, protein-ligand interactions, and structural features.
- Features: PyMOL provides a user-friendly interface with a wide range of visualization options. It allows the manipulation of molecular structures in real-time, highlighting specific regions of interest and facilitating the communication of structural insights.
- SWISS-MODEL for Protein Structure Prediction:
- Function: SWISS-MODEL is a tool for homology modeling, predicting the three-dimensional structure of a protein based on the known structure of a homologous protein.
- Application: This tool is valuable when experimental structures are not available, providing a structural framework for understanding the function and interactions of proteins.
- Features: SWISS-MODEL automates the homology modeling process, making it accessible to researchers without extensive expertise in structural biology. It integrates with various databases and offers options for model quality assessment.
- VMD (Visual Molecular Dynamics) for Molecular Dynamics Simulations:
- Function: VMD is a software package for visualizing, analyzing, and simulating the dynamics of molecular systems, particularly in the context of molecular dynamics simulations.
- Application: Molecular dynamics simulations provide insights into the motion and behavior of biomolecules over time. VMD aids researchers in analyzing trajectories, studying conformational changes, and understanding the dynamics of biological macromolecules.
- Features: VMD supports the visualization of molecular structures, trajectories, and simulation results. It is equipped with tools for measuring distances, angles, and other structural parameters during simulations, contributing to a comprehensive analysis of molecular dynamics.
These structural analysis tools are essential for researchers working in structural biology and related fields. They facilitate the visualization of molecular structures, prediction of protein structures, and exploration of molecular dynamics, ultimately aiding in the understanding of the structure-function relationships of biological macromolecules. Integrating these tools into research workflows enhances the analysis of structural data and contributes to advancements in various scientific disciplines.
C. Functional Analysis Tools
- Gene Ontology (GO) Enrichment Analysis:
- Function: Gene Ontology is a standardized vocabulary that annotates genes and gene products with terms related to biological processes, molecular functions, and cellular components. GO enrichment analysis identifies overrepresented GO terms in a set of genes.
- Application: GO enrichment analysis helps researchers understand the biological significance of a gene set, such as those differentially expressed in an experiment, by revealing the functional categories that are statistically enriched.
- Features: Tools for GO enrichment analysis, such as Enrichr and g:Profiler, allow users to input gene lists and receive information about the functional categories associated with the provided genes.
- DAVID Bioinformatics Resources:
- Function: DAVID (Database for Annotation, Visualization, and Integrated Discovery) is a bioinformatics resource that provides tools for functional annotation and analysis of gene lists. It integrates information from various databases to extract biological meaning from large gene sets.
- Application: DAVID is used for functional annotation of genes, identification of enriched functional terms, and visualization of relationships between genes in the context of biological pathways.
- Features: DAVID offers a comprehensive set of functional analysis tools, including gene functional classification, functional annotation chart, and pathway analysis. It allows users to explore the biological relevance of gene lists derived from experiments.
- KEGG (Kyoto Encyclopedia of Genes and Genomes) Pathway Analysis:
- Function: KEGG is a database that integrates information about genomes, biological pathways, diseases, and chemical substances. KEGG pathway analysis involves mapping genes to KEGG pathways to understand the functional context of gene sets.
- Application: KEGG pathway analysis helps researchers interpret the biological significance of gene sets by identifying pathways that are overrepresented or significantly associated with the genes.
- Features: Various bioinformatics tools, such as KEGG Mapper and WebGestalt, use KEGG data for pathway analysis. Researchers can visualize and explore the relationships between genes and pathways to gain insights into the functional implications of their data.
Functional analysis tools are crucial for interpreting large-scale genomic and transcriptomic data, providing insights into the biological processes and pathways associated with sets of genes. These tools play a key role in uncovering the functional context of experimental results and contribute to a deeper understanding of the molecular mechanisms underlying biological phenomena.