
Primer-BLAST-New Primer Design Tool-Tutorial
February 16, 2009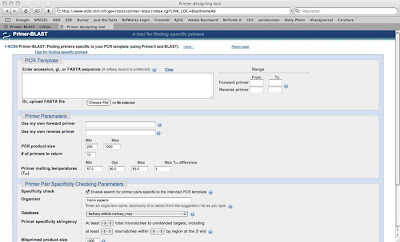 Primer-BLAST was developed at NCBI to help users make primers that are specific to the input PCR template. It uses Primer3 to design PCR primers and then submits them to BLAST search against user-selected database. The BLAST results are then automatically analyzed to avoid primer pairs that can cause amplification of targets other than the input template.
Primer-BLAST was developed at NCBI to help users make primers that are specific to the input PCR template. It uses Primer3 to design PCR primers and then submits them to BLAST search against user-selected database. The BLAST results are then automatically analyzed to avoid primer pairs that can cause amplification of targets other than the input template.
How to use Primer-BLAST
1.Go to http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi
2.Enter your FASTA sequence, an accession or a GI into the PCR Template box or you can upload fasta file
3.Provide forward prime & Reverse primer parameters
4.By default it searches human sequence database, but you can change your specific organism by typing in the organism section of primer pair specificity checking parameters
5.Use the “Get Primers” button at the bottom of the page to submit your search.
















