
Protein druggability prediction
April 19, 2007The availability of structural data, especially of proteins complexed to small molecule ligands, has enabled numerous analyses that attempt to understand and predict the forces that govern molecular recognition and ligand binding. Successful drug development requires a disease target that plays a vital role in the causation and progression of the disease phenotype and that can be modulated with a drug molecule. Binding site prediction for malate dehydrogenase (PDB: 2cmd).
Binding site prediction for malate dehydrogenase (PDB: 2cmd).
One approach for evaluating protein druggability is to analyze the genome on the basis of sequence homology to known therapeutic targets.
Another approach which rely on on the 3D structure of the protein target. Systematic analyses of protein srufaces in the search for binding pockets that have high potential to bind small drug like compuunds with high affinity.
To identify all possible binding sites on a protien surface based on algorithms broadly classified into
(i) Geometry-based
Tools based on geometry based alogirthm
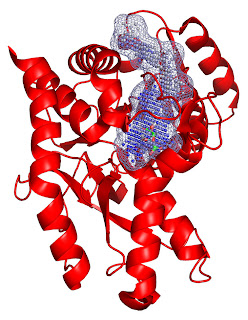 Binding site prediction for malate dehydrogenase (PDB: 2cmd).
Binding site prediction for malate dehydrogenase (PDB: 2cmd).(ii)Energy based algorithms
Tools based on Energy based algorithms
- GRID
- vdW-FT
- Drugsite
Related posts:
Career Spotlight: High Demand for Skilled Bioinformaticians
Mining Massive Biological Datasets for New Discoveries with Scalable Algorithms
Machine Learning Models are Rapidly Advancing Structure-Based Drug Design
A Deep Dive into Bioinformatics Tools and Databases for Genetic Discovery
Build Your Own Bioinformatics Software Using ChatGPT in Minutes
Quantum Computing in Bioinformatics
Exploring the Impact of Personalized Medicine on Health Outcomes
Revolutionizing Bioinformatics for Beginners: A Journey with ChatGPT
Quick Guide Tutorial: Challenges in Big Data and Bioinformatics
What are some examples of bioinformatics companies and what do they work on?
The role of bioinformatics in the development of vaccines and therapies for COVID-19
Navigating the Future of Bioinformatics: Trends, Innovations, and Key Players
