How to Use NCBI Variation Information
December 3, 2024NCBI provides a wealth of resources for exploring genetic variation, enabling researchers to investigate relationships between genetic variants and phenotypes, health conditions, and evolutionary patterns. This guide introduces you to key NCBI databases for genetic variation, including ClinVar, dbVar, dbSNP, and others, and provides an overview of how to navigate and utilize these resources effectively. Whether you’re studying human health, genomics, or population genetics, this guide will help you access and interpret the information housed in these valuable tools.
NCBI Variation Information
NCBI provides multiple database resources for information on genetic variation.
These Include: (from the NCBI Variation Resources page (http://www.ncbi.nlm.nih.gov/guide/variation/)
BioProject (formerly Genome Project)
A collection of genomics, functional genomics, and genetics studies and links to their resulting datasets. This resource describes project scope, material, and objectives and provides a mechanism to retrieve datasets that are often difficult to find due to inconsistent annotation, multiple independent submissions, and the varied nature of diverse data types which are often stored in different databases.
ClinVar
A resource to provide a public, tracked record of reported relationships between human variation and observed health status with supporting evidence. Related information in the NIH Genetic Testing Registry (GTR), MedGen, Gene, OMIM, PubMed and other sources is accessible through hyperlinks on the records.
Database of Genomic Structural Variation (dbVar)
The dbVar database has been developed to archive information associated with large scale genomic variation, including large insertions, deletions, translocations and inversions. In addition to archiving variation discovery, dbVar also stores associations of defined variants with phenotype information.
Database of Genotypes and Phenotypes (dbGaP)
An archive and distribution center for the description and results of studies which investigate the interaction of genotype and phenotype. These studies include genome-wide association (GWAS), medical resequencing, molecular diagnostic assays, as well as association between genotype and non-clinical traits.
Database of Major Histocompatibility Complex (dbMHC)
An open, publicly accessible platform where the HLA community can submit, edit, view, and exchange data related to the human major histocompatibility complex. It consists of an interactive Alignment Viewer for HLA and related genes, an MHC microsatellite database, a sequence interpretation site for Sequencing Based Typing (SBT), and a Primer/Probe database.
Database of Short Genetic Variations (dbSNP)
Includes single nucleotide variations, microsatellites, and small-scale insertions and deletions. dbSNP contains population-specific frequency and genotype data, experimental conditions, molecular context, and mapping information for both neutral variations and clinical mutations.
Genetic Testing Registry (GTR)
A voluntary registry of genetic tests and laboratories, with detailed information about the tests such as what is measured and analytic and clinical validity. GTR also is a nexus for information about genetic conditions and provides context-specific links to a variety of resources, including practice guidelines, published literature, and genetic data/information. The initial scope of GTR includes single gene tests for Mendelian disorders, as well as arrays, panels and pharmacogenetic tests.
1000 Genomes Browser
An interactive graphical viewer that allows users to explore variant calls, genotype calls and supporting evidence (such as aligned sequence reads) that have been produced by the 1000 Genomes Project.
dbSNP
dbSNP is a database of single nucleotide polymorphisms (SNPs) and multiple small-scale variations that include insertions/deletions, microsatellites, and non-polymorphic variants. (from the NCBI web site)
NCBI provides an FAQ on dbSNP at http://www.ncbi.nlm.nih.gov/books/NBK3848/
This guide will show a sample variation viewer using the LCT gene as the source.
First change the database search to SNP and create a search for the gene we are interested in and limit to the organism:
1. Choosing the first LCT variation, click on the “Varview” link:
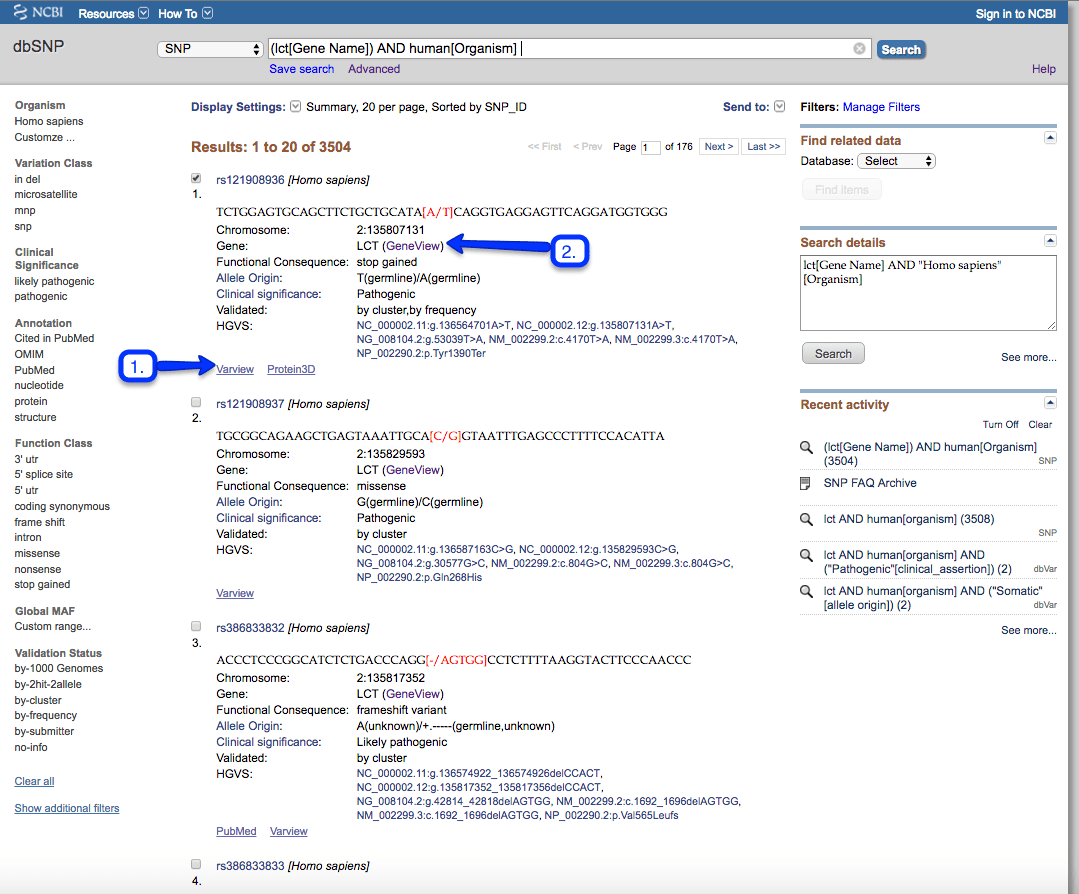
The first page of results includes many variations. By applying the filter “nonsense”, the results now show only the rs121908936 variation, which is what we have asked for above:
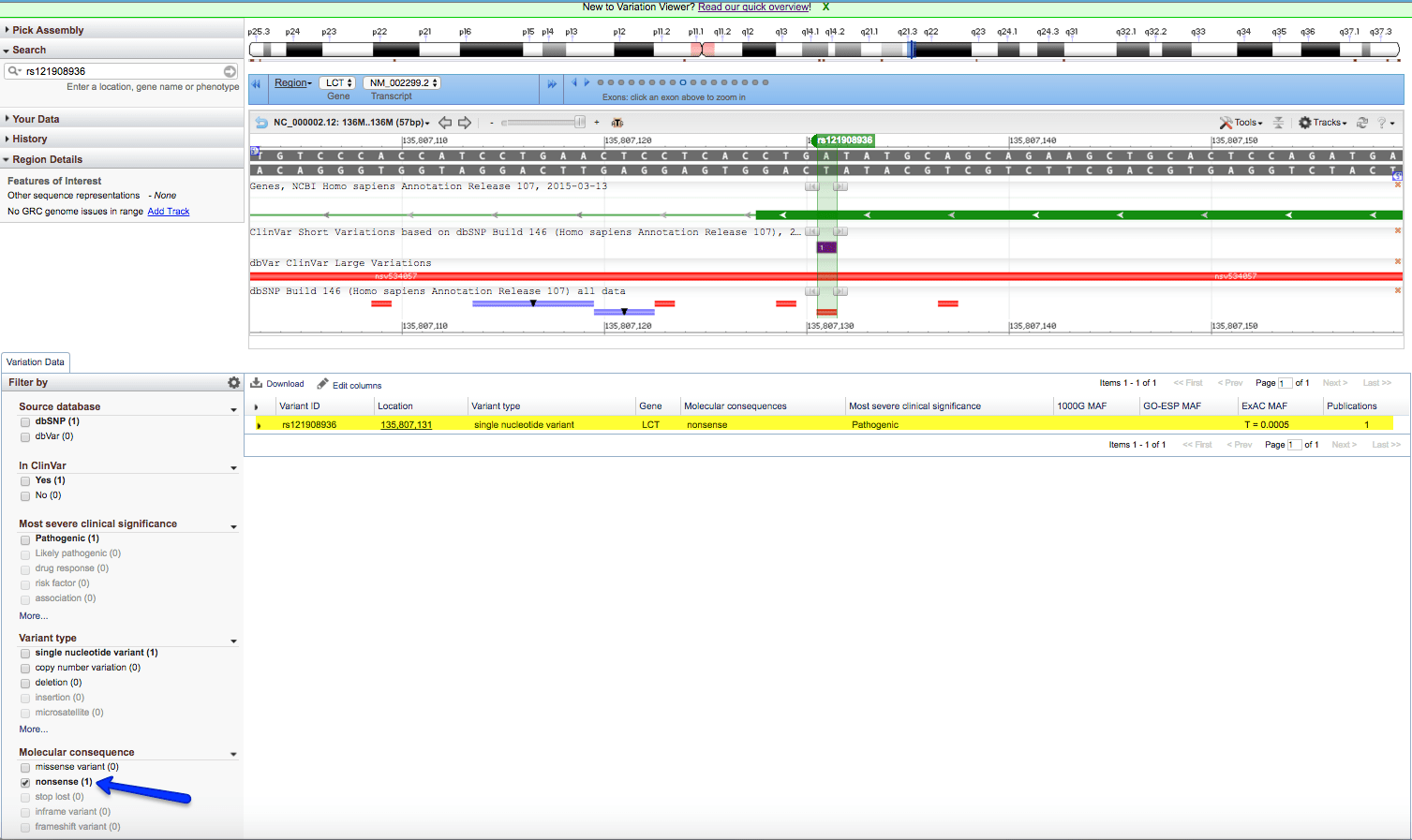
The results show that this variation is a substitution of A and T on chromosome 2 at base pair location 135,807,131.
NOTE: Whenever using any data from NCBI it is important to check the Assembly version that this record is built from. In this case, it is GRCh38.p2. When we use this variation in the 1000 Genome example, the location of this SNP shows a different location. This can be seen by clicking on the Variant ID in the above page. The results are:
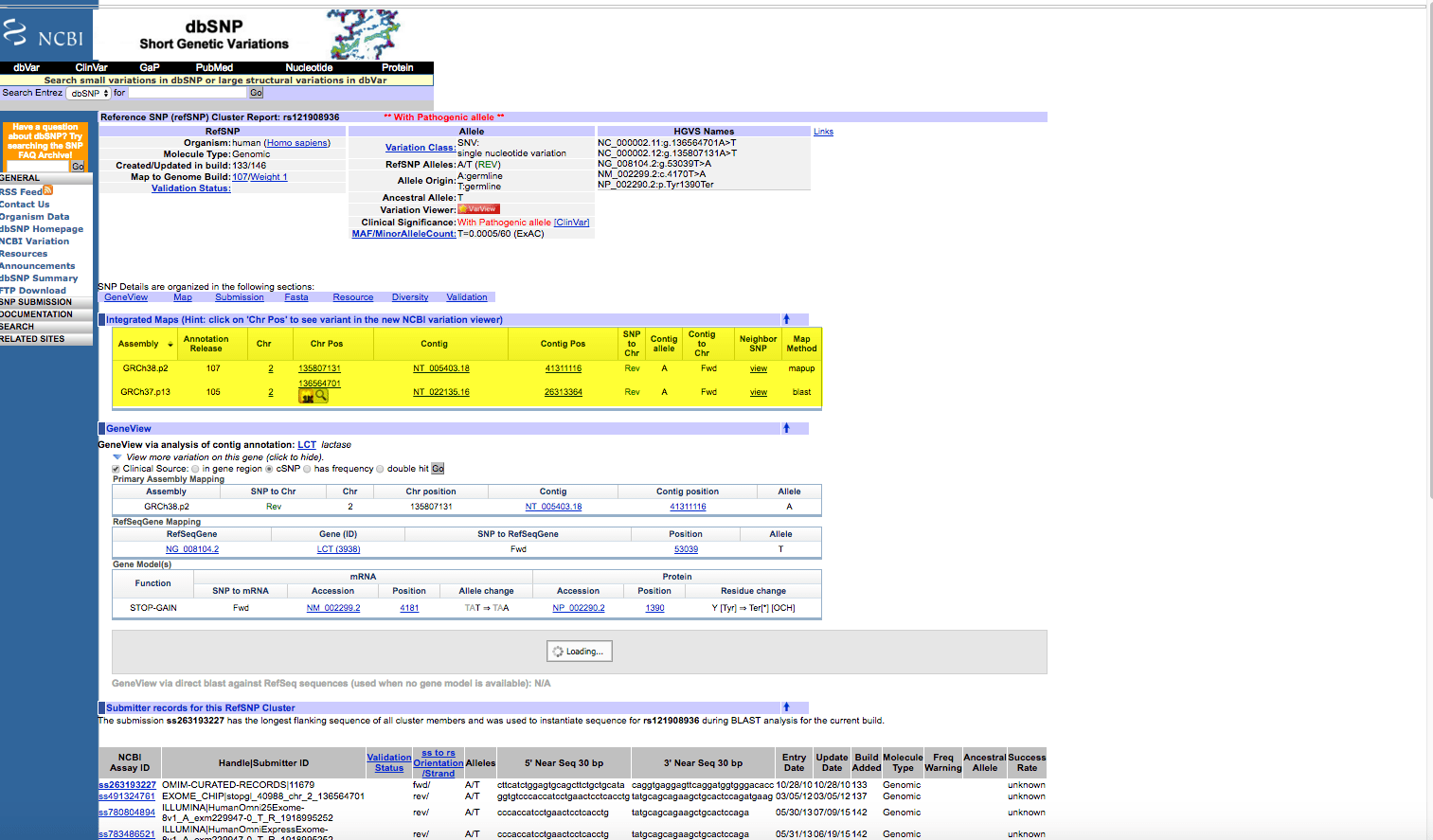
As can be seen, there is an older Assembly, GRCh37.p13. This is what the data reflects in the 1000 Genome database. This can be seen from this record by clicking on the magnifying glass icon under the Chr Pos for this chromosome position (136564701):
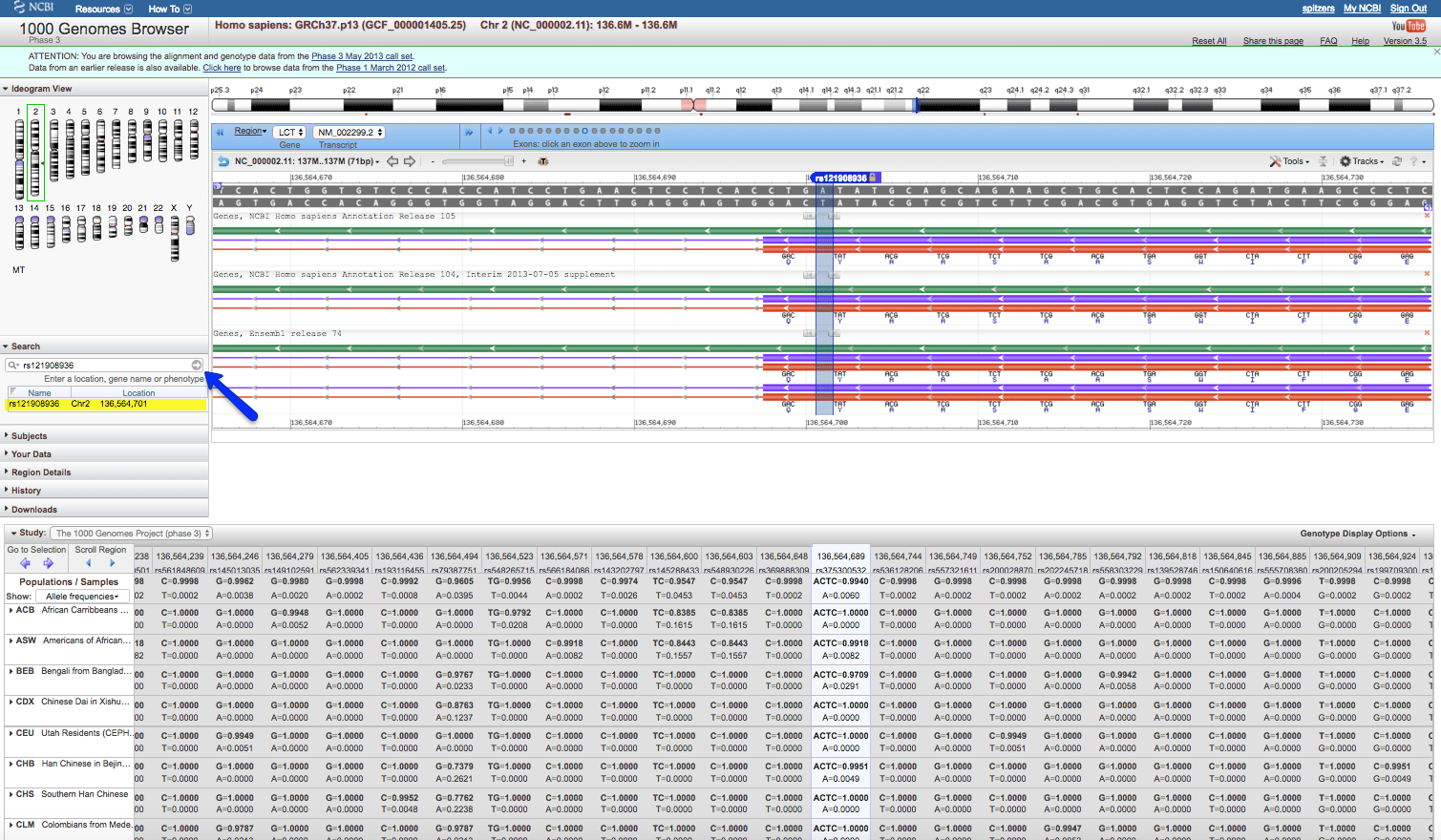
2. Next, view this variation using the Gene viewer. Click the “GeneView” link:

dbVar
dbVar is NCBI’s database of genomic structural variation – it contains insertions, deletions, duplications, inversions, multinucleotide substitutions, mobile element insertions, translocations, and complex chromosomal rearrangements. (from the NCBI web site)
NCBI provides an FAQ on dbVAR at http://www.ncbi.nlm.nih.gov/dbvar/content/help/
This guide will show a sample variation viewer using the LCT gene as the source.
First change the database search to dbVar and create a search for the gene we are interested in and limit to the organism:
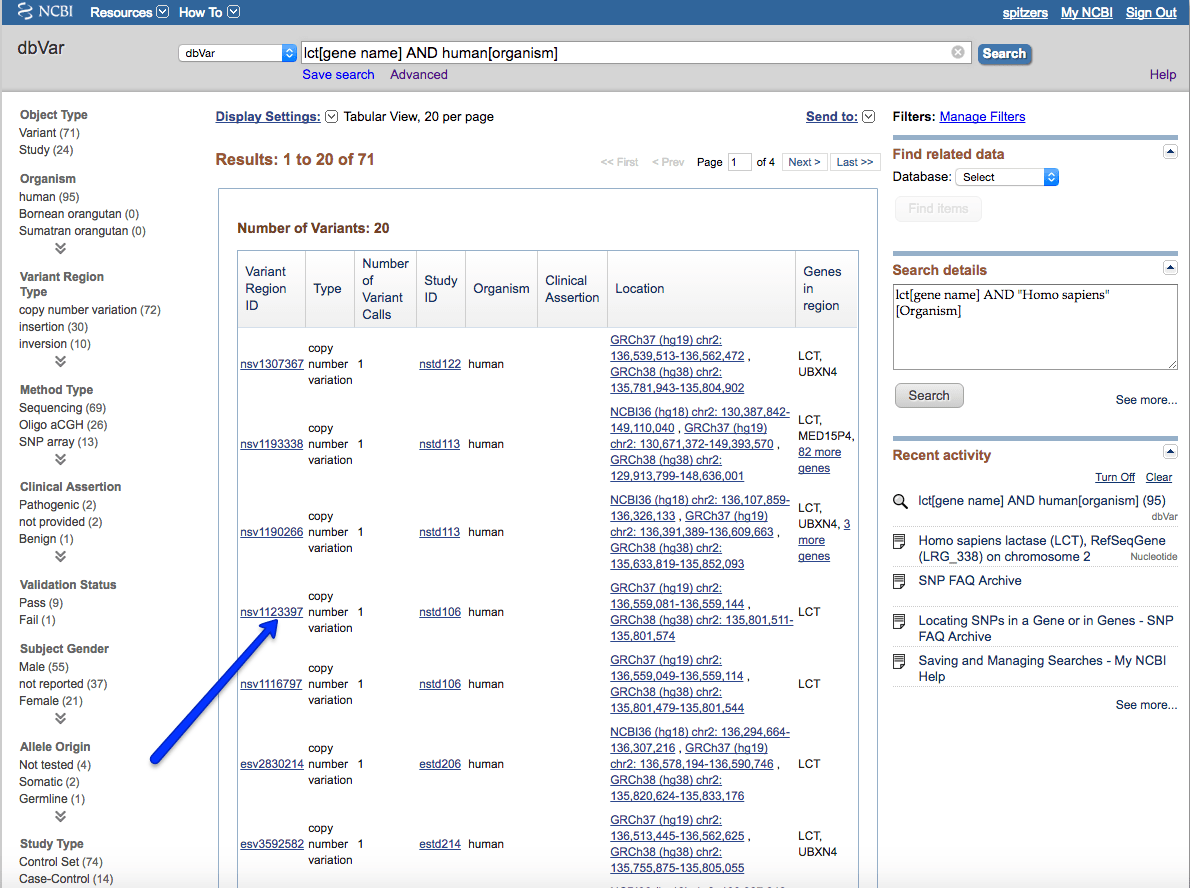
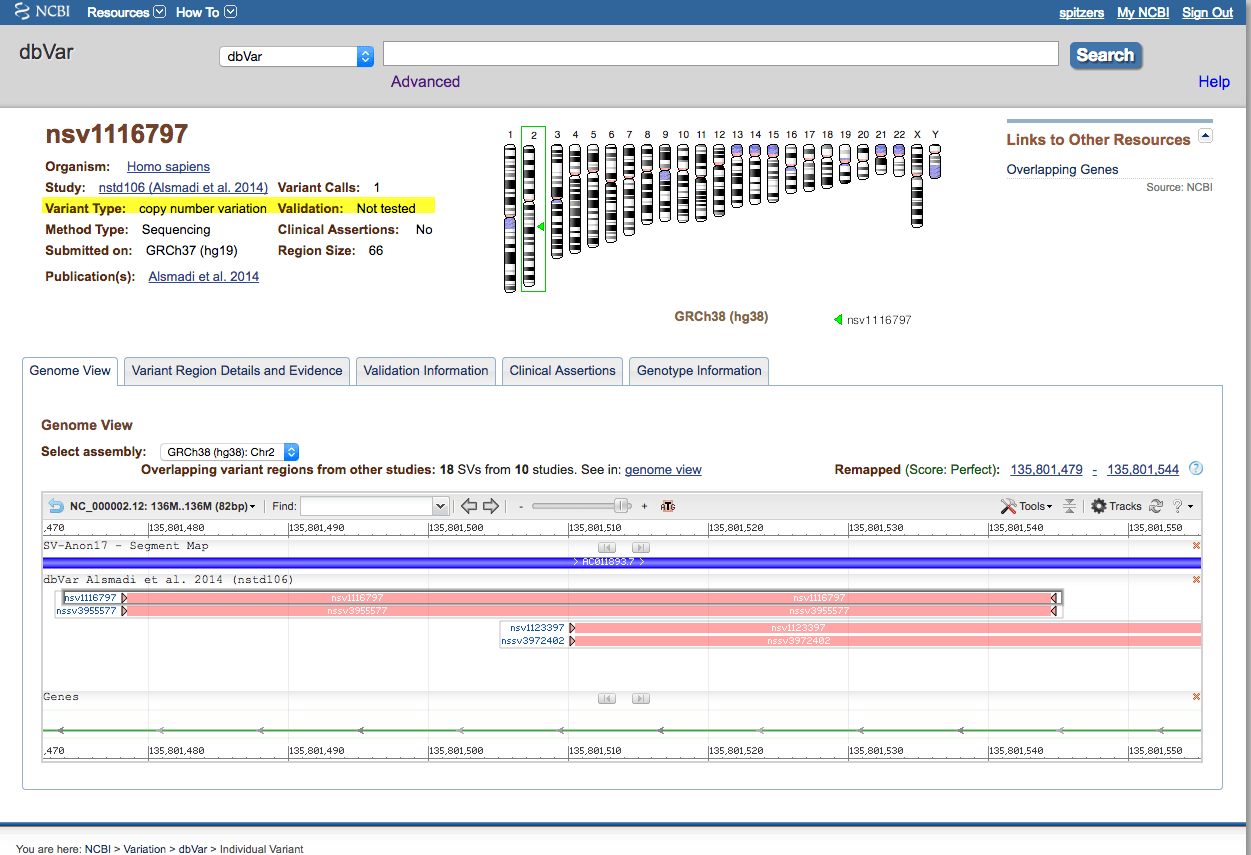
The resulting page shows larger scale variations for this this gene. Choosing one, nsv1123397, click on the link to view this variation in more detail:
ClinVar
NCBI ClinVar
ClinVar aggregates information about genomic variation and its relationship to human health. (from NCBI website)
This resource can be accessed at http://www.ncbi.nlm.nih.gov/clinvar
NCBI has an Introduction to this resource at http://www.ncbi.nlm.nih.gov/clinvar/intro/
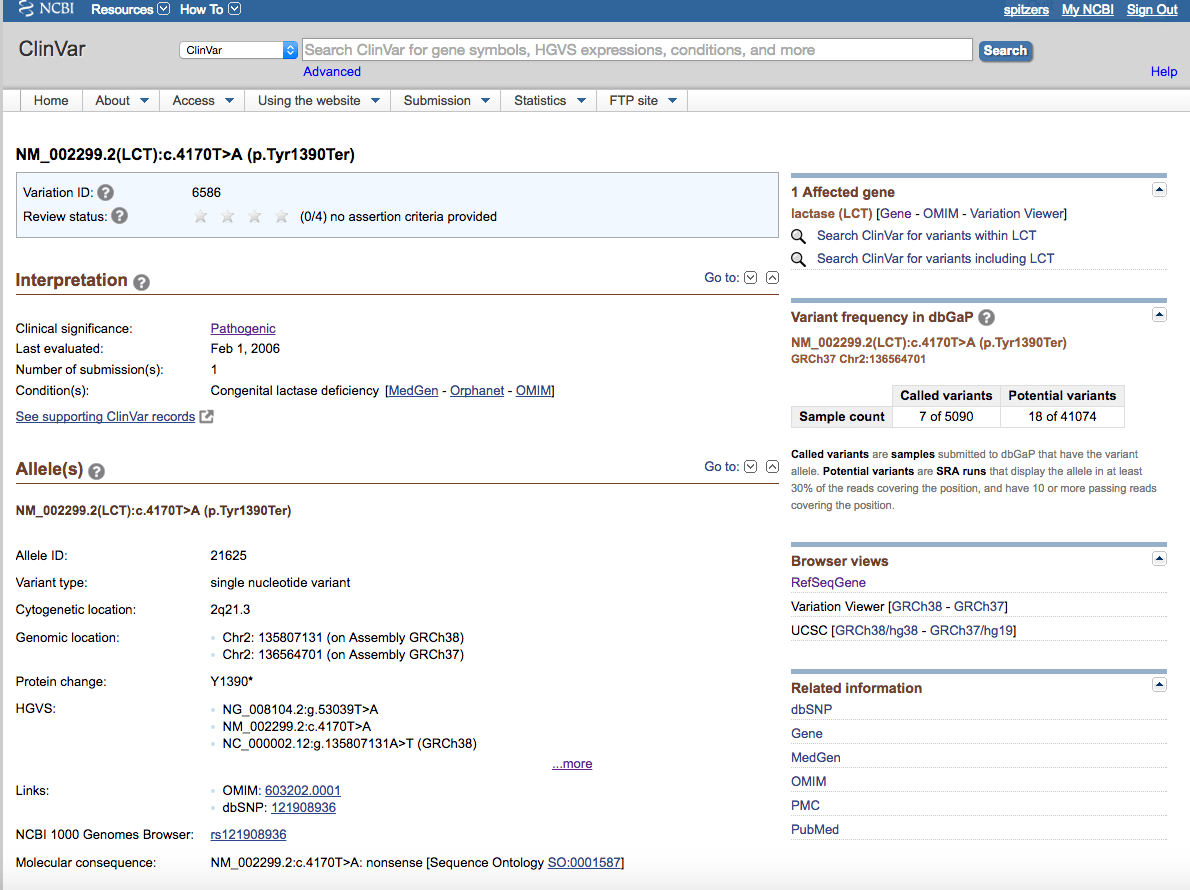
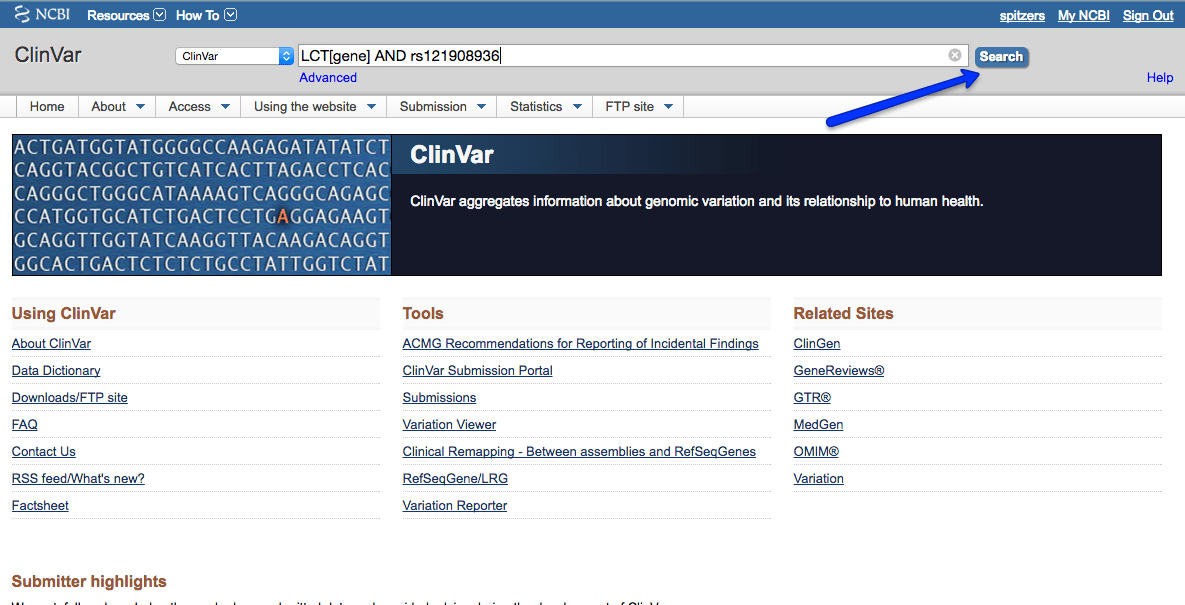
This example will step through looking for the same LRC variation that we looked at in the dbSNP guide – rs121908936.
Search for LCT and rs121908936:
Which returns more information on this allele: