Advanced Topics in Genetics for Bioinformatics
March 31, 2024Genes, Chromosomes, and Heredity
Gene as the fundamental unit of heredity
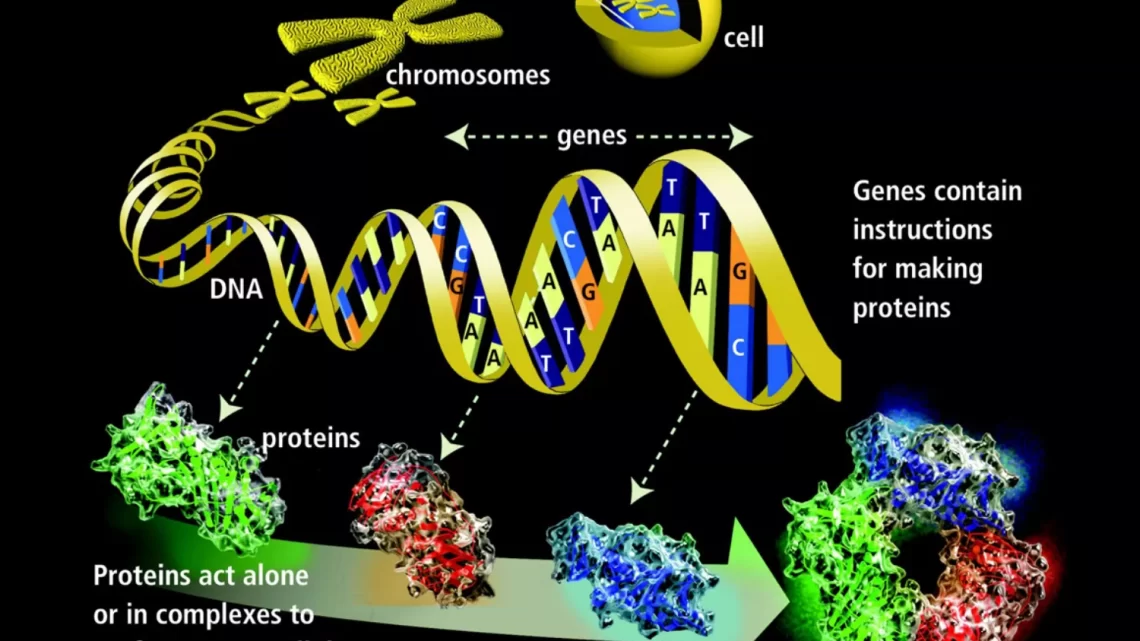
A gene is the fundamental unit of heredity. It carries the information that determines a specific trait or function in an organism. Genes are composed of DNA and are located on chromosomes. They serve as instructions for making proteins, which are essential for the structure, function, and regulation of the body’s tissues and organs. Through processes like replication, transcription, and translation, genes play a crucial role in passing on genetic information from one generation to the next.
- Definition: A gene is a segment of DNA that contains the instructions for building a specific protein or RNA molecule. These molecules perform various functions in the body, such as building tissues, regulating chemical processes, and carrying signals between cells.
- Structure: Genes are composed of nucleotides, which are the building blocks of DNA. Each nucleotide consists of a sugar molecule (deoxyribose), a phosphate group, and one of four nitrogenous bases: adenine (A), cytosine (C), guanine (G), or thymine (T). The sequence of these bases in a gene determines the genetic code, which specifies the order in which amino acids are assembled to create a protein.
- Chromosomal Location: Genes are located on chromosomes, which are thread-like structures made of DNA and proteins. In humans, each cell typically contains 46 chromosomes organized into 23 pairs. One chromosome in each pair comes from the mother, and the other comes from the father. Genes can be found on both autosomes (non-sex chromosomes) and sex chromosomes (X and Y in humans).
- Function: Genes play a crucial role in determining the traits of an organism. They are responsible for inherited characteristics such as eye color, blood type, and susceptibility to certain diseases. Genes also play a role in regulating the expression of other genes, influencing how traits are expressed.
- Inheritance: Genes are passed from parents to offspring through reproduction. Offspring inherit one set of genes from each parent, which is why children often share traits with their parents. The process of passing genes from parents to offspring is called heredity.
- Gene Expression: Gene expression is the process by which the information stored in a gene is used to create a functional product, such as a protein or RNA molecule. This process involves several steps, including transcription (copying the gene’s DNA sequence into RNA) and translation (using the RNA to assemble amino acids into a protein).
- Variation and Evolution: Genes are subject to mutations, which are changes in the DNA sequence. Mutations can lead to variations in traits within a population. Over time, these variations can accumulate and contribute to evolution, the process by which species change over time.
In summary, genes are the fundamental units of heredity, containing the instructions for building proteins and determining traits in organisms. They are located on chromosomes and are passed from parents to offspring through reproduction. Genes play a crucial role in evolution, genetic diversity, and the functioning of living organisms.
Chromosome structure, function, and molecular organization
Chromosomes are complex structures found in the nucleus of eukaryotic cells that play a crucial role in storing and organizing genetic information. They are composed of DNA, proteins, and small RNA molecules. Let’s explore their structure, function, and molecular organization in more detail:
- Structure:
- DNA: The primary component of chromosomes is DNA, which is a double-stranded molecule that carries the genetic code. DNA is made up of nucleotide units, each consisting of a sugar-phosphate backbone and one of four nitrogenous bases: adenine (A), thymine (T), cytosine (C), and guanine (G).
- Histone Proteins: DNA is wrapped around histone proteins to form a complex called chromatin. Histones help organize and compact the DNA into a more condensed structure, allowing it to fit within the nucleus.
- Nucleosomes: The basic unit of chromatin structure is the nucleosome, which consists of DNA wrapped around a core of eight histone proteins. Nucleosomes are connected by linker DNA and additional histone proteins.
- Function:
- Genetic Information Storage: Chromosomes store genetic information in the form of genes, which are specific sequences of DNA that code for proteins or functional RNA molecules.
- Cell Division: Chromosomes play a crucial role in cell division, particularly in mitosis and meiosis. During these processes, chromosomes condense, become visible under a microscope, and are distributed to daughter cells to ensure each cell receives the correct amount of genetic material.
- Gene Regulation: Chromosomes are involved in regulating gene expression, determining which genes are turned on or off in a particular cell type or under specific conditions.
- Structural Support: Chromosomes provide structural support to the nucleus and help organize the DNA within the nucleus.
- Molecular Organization:
- Centromere: The centromere is a specialized region of a chromosome that plays a role in chromosome segregation during cell division. It serves as the attachment point for the spindle fibers that pull the chromosomes apart.
- Telomeres: Telomeres are regions of repetitive DNA sequences at the ends of chromosomes. They protect the ends of the chromosomes from deterioration and are involved in regulating the lifespan of a cell.
- Bandings: Chromosomes can be stained to reveal characteristic banding patterns, which are used to identify and distinguish between chromosomes. These banding patterns are based on differences in the DNA sequence along the length of the chromosome.
In summary, chromosomes are structures composed of DNA and proteins that store and organize genetic information in the nucleus of eukaryotic cells. They play a crucial role in cell division, gene regulation, and maintaining the structural integrity of the cell.
DNA as the genetic material
DNA (deoxyribonucleic acid) is the genetic material found in most living organisms, including humans. It carries the genetic instructions that are essential for the growth, development, functioning, and reproduction of all organisms. Here’s a detailed explanation of DNA as the genetic material:
- Chemical Structure: DNA is a long, double-stranded molecule made up of nucleotides. Each nucleotide consists of a sugar molecule (deoxyribose), a phosphate group, and one of four nitrogenous bases: adenine (A), thymine (T), cytosine (C), and guanine (G). The two strands of DNA are held together by hydrogen bonds between complementary base pairs (A with T, and C with G).
- Function: DNA carries the genetic information that determines the traits of an organism. This information is encoded in the sequence of nucleotide bases along the DNA molecule. Genes are specific sequences of DNA that contain instructions for making proteins, which are the building blocks of cells and play a variety of roles in the body.
- Replication: Before a cell divides, its DNA must be replicated so that each new cell receives a complete set of genetic instructions. DNA replication is a complex process that involves the unwinding of the DNA molecule, the synthesis of new complementary strands, and the proofreading and correction of errors.
- Transcription: The genetic information encoded in DNA is used to synthesize RNA molecules in a process called transcription. RNA is a single-stranded molecule that is similar in structure to DNA but contains the base uracil (U) instead of thymine (T). Messenger RNA (mRNA) carries the genetic information from DNA to the ribosomes, where it is used to direct the synthesis of proteins in a process called translation.
- Genetic Code: The sequence of nucleotide bases in DNA forms a genetic code that determines the sequence of amino acids in proteins. The genetic code is universal, meaning that the same sequence of bases codes for the same amino acid in all living organisms.
- Mutations: Changes in the DNA sequence, known as mutations, can alter the genetic information carried by an organism. Some mutations can be harmful, leading to genetic disorders or diseases, while others may have no effect or could be beneficial in certain circumstances.
In summary, DNA is the genetic material that carries the instructions for the development, functioning, and reproduction of living organisms. Its structure and function are essential for the transmission of genetic information from one generation to the next and for the diversity of life on Earth.
Cell Division and Cell Cycle
Mitosis and Meiosis
Mitosis and meiosis are two types of cell division processes that play critical roles in the growth, development, and reproduction of organisms. Here’s an overview of each process:
Mitosis:
- Purpose: Mitosis is a type of cell division that produces two daughter cells that are genetically identical to the parent cell. It is responsible for growth, repair, and maintenance of multicellular organisms.
- Stages: Mitosis is divided into four stages: prophase, metaphase, anaphase, and telophase.
- Prophase: Chromosomes condense, becoming visible under a microscope. The nuclear envelope breaks down, and the spindle apparatus forms.
- Metaphase: Chromosomes line up along the metaphase plate, a plane that divides the cell into two halves.
- Anaphase: Sister chromatids separate and move to opposite poles of the cell.
- Telophase: Chromatids reach the poles, and new nuclear envelopes form around the separated chromatids, forming two daughter nuclei.
- Cytokinesis: Following telophase, the cell undergoes cytokinesis, during which the cytoplasm divides, and two daughter cells are formed.
Meiosis:
- Purpose: Meiosis is a type of cell division that produces gametes (sperm and egg cells) with half the number of chromosomes as the parent cell. It is essential for sexual reproduction and contributes to genetic diversity.
- Stages: Meiosis consists of two rounds of division, known as meiosis I and meiosis II, each with prophase, metaphase, anaphase, and telophase stages.
- Meiosis I: Homologous chromosomes pair up and exchange genetic material in a process called crossing over. The homologous chromosomes separate, reducing the chromosome number by half.
- Meiosis II: Similar to mitosis, the sister chromatids separate, resulting in four haploid daughter cells, each with a unique combination of genetic material.
- Genetic Variation: Meiosis generates genetic variation through independent assortment of chromosomes (random alignment of homologous pairs) and crossing over (exchange of genetic material between homologous chromosomes).
In summary, mitosis produces two identical daughter cells for growth and repair, while meiosis produces four genetically unique daughter cells (gametes) for sexual reproduction. Both processes are essential for the survival and reproduction of organisms and contribute to genetic diversity within populations.
Genetic consequences of the cell cycle
The cell cycle is the series of events that take place in a cell leading to its division and duplication of its DNA to produce two daughter cells. The cell cycle consists of interphase (G1, S, and G2 phases) and mitotic phase (M phase, which includes mitosis and cytokinesis). The cell cycle has several genetic consequences, including:
- DNA Replication Errors: During the S phase of interphase, DNA is replicated to ensure that each daughter cell receives a complete set of genetic information. Errors in DNA replication, such as base substitutions, insertions, or deletions, can lead to mutations. Mutations can alter gene function, leading to genetic disorders or diseases.
- Mutations in Cell Cycle Regulation Genes: Genes that regulate the cell cycle, such as tumor suppressor genes and oncogenes, can become mutated. Mutations in these genes can disrupt the normal control of the cell cycle, leading to uncontrolled cell division and cancer.
- Chromosomal Abnormalities: Errors during mitosis, such as nondisjunction (failure of chromosomes to separate properly), can result in chromosomal abnormalities. Examples include aneuploidy (an abnormal number of chromosomes) and chromosomal translocations (movement of chromosome segments to different chromosomes). These abnormalities can lead to genetic disorders or cancer.
- Genetic Diversity: Meiosis, which is a part of the cell cycle in sexually reproducing organisms, generates genetic diversity through independent assortment of chromosomes and crossing over. This genetic diversity contributes to the evolution and adaptation of populations to changing environments.
- Cell Differentiation: The cell cycle is also involved in cell differentiation, where cells become specialized for specific functions. This process involves the activation and inactivation of specific genes, leading to differences in gene expression and cellular function.
In summary, the cell cycle has several genetic consequences, including DNA replication errors, mutations in cell cycle regulation genes, chromosomal abnormalities, genetic diversity, and cell differentiation. These consequences play critical roles in development, evolution, and the maintenance of genetic integrity in organisms.
Chromosome Mutations
Types of mutations
Mutations are changes in the DNA sequence of an organism. They can occur in various ways and can have different effects on the organism. Here are some common types of mutations:
- Point Mutations:
- Substitution: One nucleotide is replaced by another. This can be:
- Silent: The change does not result in a change to the amino acid sequence due to the degeneracy of the genetic code.
- Missense: The change results in a different amino acid being incorporated into the protein, which can alter its function.
- Nonsense: The change results in a premature stop codon, leading to a truncated (shortened) protein.
- Substitution: One nucleotide is replaced by another. This can be:
- Insertions and Deletions (Indels):
- Insertion: One or more nucleotides are added to the DNA sequence, shifting the reading frame and potentially altering the entire amino acid sequence downstream.
- Deletion: One or more nucleotides are removed from the DNA sequence, also shifting the reading frame and potentially altering the entire amino acid sequence downstream.
- Repeat Expansions:
- In this type of mutation, a sequence of nucleotides is repeated multiple times. Expansion of these repeats can lead to various genetic disorders.
- Frameshift Mutations:
- Insertions or deletions that are not multiples of three nucleotides can disrupt the reading frame of the gene, leading to a completely different amino acid sequence downstream of the mutation.
- Chromosomal Rearrangements:
- Translocation: A segment of one chromosome breaks off and attaches to another chromosome.
- Inversion: A segment of a chromosome breaks off, flips around, and reattaches in the reverse orientation.
- Duplication: A segment of a chromosome is duplicated, leading to extra copies of genes.
- Deletion: A segment of a chromosome is lost.
- Mutations in Regulatory Regions:
- Mutations in non-coding regions of DNA, such as promoters or enhancers, can affect gene expression by altering the binding of transcription factors or other regulatory proteins.
- Splice Site Mutations:
- Mutations that affect the splicing of pre-mRNA can lead to the inclusion of intronic sequences in the mature mRNA, resulting in a non-functional protein.
These mutations can have a range of effects, from no discernible impact to severe consequences, depending on their location, type, and the specific genes involved.
Numerical changes in chromosomes
Numerical changes in chromosomes refer to alterations in the number of chromosomes in a cell. There are two main types of numerical changes:
- Euploidy: Euploidy refers to the presence of a complete set of chromosomes. In humans, a normal euploid cell contains 46 chromosomes, with 22 pairs of autosomes and 1 pair of sex chromosomes (XX in females and XY in males). Euploidy can be further classified into two types:
- Haploid (n): A cell containing only one set of chromosomes. In humans, gametes (sperm and egg cells) are haploid, containing 23 chromosomes.
- Diploid (2n): A cell containing two sets of chromosomes, one from each parent. Most cells in the human body are diploid, with 46 chromosomes.
- Aneuploidy: Aneuploidy refers to the presence of an abnormal number of chromosomes, resulting from the gain or loss of individual chromosomes. Aneuploidy can occur in any chromosome, and it is commonly associated with genetic disorders. Some examples of aneuploidies include:
- Trisomy: The presence of an extra chromosome, resulting in three copies of that chromosome. Down syndrome (trisomy 21) is an example of a trisomy disorder.
- Monosomy: The absence of one chromosome, resulting in only one copy of that chromosome. Turner syndrome (monosomy X) is an example of a monosomy disorder.
- Polysomy: The presence of more than the normal number of chromosomes. For example, Klinefelter syndrome is a condition in which males have an extra X chromosome (XXY).
Aneuploidy can occur due to errors in cell division, such as nondisjunction, where chromosomes fail to separate properly during meiosis or mitosis. The consequences of aneuploidy can vary depending on the specific chromosome involved and the number of extra or missing chromosomes. Aneuploidy is a common cause of miscarriages and can lead to developmental abnormalities and genetic disorders in individuals who survive.
Structural changes in chromosomes
Structural changes in chromosomes involve alterations in the structure of individual chromosomes, often affecting the arrangement or number of genes. Here are the main types of structural changes:
- Duplications: Duplications occur when a segment of a chromosome is duplicated, resulting in an extra copy of that segment. Duplications can range in size from small gene duplications to duplications of entire chromosome arms. Duplications can lead to genetic disorders if they disrupt gene function or regulation.
- Deletions: Deletions occur when a segment of a chromosome is lost or deleted. Deletions can vary in size and can involve a single nucleotide, a gene, or a larger segment of a chromosome. Deletions can lead to genetic disorders if they remove critical genes or regulatory elements.
- Inversions: Inversions occur when a segment of a chromosome is reversed in orientation within the chromosome. There are two main types of inversions: pericentric inversions, which include the centromere, and paracentric inversions, which do not include the centromere. Inversions can lead to genetic disorders if they disrupt gene function or regulation.
- Translocations: Translocations occur when a segment of one chromosome breaks off and attaches to another chromosome. There are two main types of translocations: reciprocal translocations, where two non-homologous chromosomes exchange segments, and Robertsonian translocations, where two acrocentric chromosomes fuse at the centromere. Translocations can lead to genetic disorders if they disrupt gene function or regulation, or if they result in the formation of novel fusion genes.
These structural changes can occur spontaneously or be inherited from a parent. They can lead to a variety of genetic disorders, depending on the specific genes involved and the size and location of the structural change. Understanding these structural changes is important for diagnosing genetic disorders and developing appropriate treatment strategies.
Linkage, Crossing Over, and Chromosome Mapping
Basic principles and exceptions to Mendelian law of independent assortment
The Mendelian law of independent assortment states that alleles of different genes segregate independently of each other during the formation of gametes. This means that the inheritance of one gene does not influence the inheritance of another gene. While this principle generally holds true, there are some exceptions:
- Linked Genes: Genes that are located close to each other on the same chromosome tend to be inherited together and do not assort independently. This is because they are physically linked and are more likely to be inherited as a unit. However, crossing over during meiosis can result in the exchange of genetic material between linked genes, leading to recombination and some degree of independent assortment.
- Incomplete Dominance: In cases of incomplete dominance, the heterozygous phenotype is intermediate between the two homozygous phenotypes. For example, in snapdragons, a cross between a red-flowered plant (RR) and a white-flowered plant (WW) results in pink-flowered offspring (RW). This does not strictly follow the Mendelian law of independent assortment because the alleles for flower color are not segregating independently.
- Codominance: Codominance occurs when both alleles in a heterozygote are expressed fully, leading to a phenotype that shows both traits. An example is the AB blood type in humans, where both A and B alleles are expressed on the surface of red blood cells. This is an exception to the Mendelian law because both alleles are expressed, rather than one being dominant over the other.
- Multiple Alleles: Some genes have more than two alleles in a population. For example, the ABO blood group system in humans has three alleles: IA, IB, and i. The IA and IB alleles are codominant, while the i allele is recessive. This complexity does not follow simple Mendelian inheritance patterns.
- Pleiotropy: Some genes can have multiple effects on an organism’s phenotype. For example, a single gene in humans can affect multiple traits such as skin color, eye color, and susceptibility to certain diseases. This violates the principle of independent assortment because a single gene is influencing multiple phenotypic traits.
- Epistasis: Epistasis occurs when the expression of one gene masks or modifies the expression of another gene. This can result in complex inheritance patterns that do not follow Mendelian principles. An example is coat color in mice, where the interaction between two genes determines the final coat color phenotype.
While these exceptions exist, Mendel’s laws of segregation and independent assortment provide a foundation for understanding the inheritance of genetic traits in many organisms.
Linkage intensity: calculating recombination frequency, coupling and repulsion linkages
Linkage intensity refers to the strength of the linkage between two genes on the same chromosome. It is determined by the recombination frequency between the two genes. Recombination frequency is the proportion of offspring that show a recombinant phenotype (resulting from crossing over between homologous chromosomes) out of the total number of offspring.
To calculate recombination frequency, you can use the formula:
Recombination frequency=Number of recombinant offspringTotal number of offspring×100%
Coupling linkage (cis configuration) and repulsion linkage (trans configuration) refer to the arrangement of alleles on homologous chromosomes. In coupling linkage, the dominant alleles of two genes are present on one chromosome, while the recessive alleles are on the other chromosome. In repulsion linkage, each chromosome carries one dominant and one recessive allele.
To determine if genes are in coupling or repulsion linkage, you can examine the phenotypes of the offspring. If the phenotypes match the dominant-recessive order of the parental alleles, the genes are in coupling linkage. If the phenotypes are different from the dominant-recessive order, the genes are in repulsion linkage.
In summary, linkage intensity is determined by the recombination frequency between two genes, and coupling and repulsion linkages refer to the arrangement of alleles on homologous chromosomes. These concepts are important in understanding the inheritance patterns of genes located on the same chromosome.
Crossing over as the physical basis of recombination
Crossing over is the physical process that underlies recombination, the exchange of genetic material between homologous chromosomes. During crossing over, corresponding segments of chromatids from two homologous chromosomes break and exchange places, resulting in the recombination of genes.
The process of crossing over occurs during meiosis, specifically during prophase I. It involves the following steps:
- Synapsis: Homologous chromosomes pair up to form bivalents (also known as tetrads), where each chromosome consists of two chromatids. This pairing is facilitated by the formation of the synaptonemal complex.
- Crossing Over: During synapsis, breaks occur in the DNA molecules of the chromatids. The broken ends of chromatids from one chromosome exchange places with the corresponding broken ends of chromatids from the other chromosome. This exchange of genetic material is known as crossing over.
- Chiasma Formation: The points where crossing over occurs are called chiasmata. Chiasmata physically link the homologous chromosomes and are visible under a microscope.
- Disjunction: After crossing over, the homologous chromosomes remain attached at the chiasmata. As meiosis progresses, the homologous chromosomes separate (disjoin) and move to opposite poles of the cell during anaphase I.
- Recombination of Genes: Crossing over results in the recombination of genes between homologous chromosomes. This leads to the formation of genetically unique gametes with combinations of alleles that differ from those of the parents.
Crossing over is essential for genetic diversity as it creates new combinations of alleles in offspring. It also helps to ensure the proper segregation of homologous chromosomes during meiosis by physically linking them until they are ready to separate.
Gene mapping and recombination frequencies
Gene mapping is the process of determining the relative positions of genes on a chromosome and the distances between them. One of the key components of gene mapping is the measurement of recombination frequencies, which is the frequency at which two genes are separated from each other due to crossing over during meiosis.
Recombination frequencies are measured using data from genetic crosses, where the offspring’s phenotypes are used to infer the genotypes of the parents. The formula for calculating recombination frequency is:
Recombination frequency=Number of recombinant offspringTotal number of offspring×100%
The unit of measurement for recombination frequencies is centimorgans (cM), named after the geneticist Thomas Hunt Morgan. One centimorgan is equal to a 1% recombination frequency between two genes.
Gene mapping based on recombination frequencies can provide information about the relative positions of genes on a chromosome. Genes that are far apart on a chromosome are more likely to undergo crossing over and have a higher recombination frequency, while genes that are close together are less likely to undergo crossing over and have a lower recombination frequency.
By measuring recombination frequencies between different pairs of genes, geneticists can construct a genetic map, also known as a linkage map, which shows the order and relative distances between genes on a chromosome. Genetic maps are important tools for understanding the organization of genes in genomes and for studying the inheritance of genetic traits.
Sex Determination and Sex-Linked Characteristics
Chromosomal sex-determining systems
Chromosomal sex-determining systems are mechanisms by which an organism’s sex is determined based on the presence or absence of certain chromosomes. There are several chromosomal sex-determining systems found in different species:
- XX/XY System (Mammals, including Humans):
- In this system, females have two X chromosomes (XX), while males have one X and one Y chromosome (XY).
- The presence of the Y chromosome triggers male development, while its absence results in female development.
- ZZ/ZW System (Birds, Some Reptiles, Some Fish, Some Insects):
- In this system, males have two of the same kind of sex chromosome (ZZ), while females have two different sex chromosomes (ZW).
- The presence of the Z chromosome triggers male development, while the presence of the W chromosome results in female development.
- X0 System (Some Insects, including Grasshoppers):
- In this system, females have two X chromosomes, while males have only one X chromosome (X0).
- The presence of the X chromosome determines female development, while its absence results in male development.
- Haplo-diploid System (Some Insects, including Bees and Ants):
- In this system, males develop from unfertilized eggs and are haploid (n), meaning they have half the usual number of chromosomes.
- Females develop from fertilized eggs and are diploid (2n), meaning they have the usual number of chromosomes.
- The determination of male or female offspring is not based on sex chromosomes but on whether the egg is fertilized.
- Environmental Sex Determination (Some Reptiles, Fish, and Amphibians):
- In some species, environmental factors such as temperature can determine the sex of offspring, rather than genetic factors.
- For example, in some reptiles, higher temperatures during egg incubation can result in more males, while lower temperatures can result in more females.
These chromosomal sex-determining systems illustrate the diversity of mechanisms by which sex is determined in different species. While the XX/XY system is most familiar in mammals, other systems demonstrate alternative evolutionary solutions to the question of sex determination.
Genic and environmental sex determination
Genic and environmental sex determination are two mechanisms by which an organism’s sex is determined. These mechanisms can vary among different species and can sometimes interact with each other.
- Genic Sex Determination:
- Definition: Genic sex determination, also known as genetic sex determination, is the process by which an organism’s sex is determined by the genetic makeup of its sex chromosomes.
- Examples: The XX/XY system in mammals, where females have two X chromosomes (XX) and males have one X and one Y chromosome (XY), is a classic example of genic sex determination. Other systems, such as the ZZ/ZW system in birds and some reptiles, where males have two of the same sex chromosome (ZZ) and females have two different sex chromosomes (ZW), are also examples of genic sex determination.
- Environmental Sex Determination:
- Definition: Environmental sex determination (ESD) is the process by which an organism’s sex is determined by environmental factors, such as temperature, rather than its genetic makeup.
- Examples: In some reptiles, such as turtles and crocodilians, the temperature at which the eggs are incubated can determine the sex of the offspring. For example, higher temperatures during incubation may result in more males, while lower temperatures may result in more females. Some fish and amphibians also exhibit environmental sex determination.
- Interaction Between Genic and Environmental Sex Determination:
- In some species, both genetic and environmental factors can influence sex determination. For example, in some reptiles, such as the red-eared slider turtle, genotypic sex determination (based on the presence of specific sex chromosomes) is the primary mechanism, but environmental factors can also play a role in determining sex.
- In some cases, environmental factors may override genetic sex determination. For example, in the Australian skink lizard, genotypic sex determination is the primary mechanism, but if eggs are incubated at extreme temperatures, the sex ratio can be skewed.
Genic and environmental sex determination illustrate the complexity of sex determination mechanisms in different species. While genic sex determination is more common in mammals and birds, environmental sex determination is observed in various reptiles, fish, and amphibians. In some species, both mechanisms can interact to determine the sex of offspring.
Sex determination in various organisms
Sex determination mechanisms vary widely among different organisms, including mammals, birds, reptiles, fish, insects, and plants. Here’s a brief overview of sex determination in some of these groups:
- Mammals (including Humans):
- Mammals typically use genic sex determination. In humans, females have two X chromosomes (XX), and males have one X and one Y chromosome (XY). The SRY gene on the Y chromosome triggers male development.
- Birds:
- Birds use a ZW/ZZ chromosomal sex determination system. Females have two different sex chromosomes (ZW), and males have two of the same sex chromosomes (ZZ). The Z chromosome determines male development.
- Reptiles:
- Reptiles exhibit a variety of sex determination mechanisms. Some species, like crocodilians and turtles, use temperature-dependent sex determination (TSD), where the temperature at which the eggs are incubated determines the sex of the offspring. Others, like some snakes and lizards, use genotypic sex determination (GSD) similar to mammals, with sex chromosomes determining sex.
- Fish:
- Fish also show diverse sex determination mechanisms. Some species have genotypic sex determination with XX/XY chromosomes, similar to mammals. Others have environmental sex determination, where environmental factors such as temperature, social factors, or even size can influence sex determination.
- Insects:
- Insects exhibit various sex determination mechanisms. Some use genic sex determination with sex chromosomes, while others, like bees and ants, have a haplo-diploid system where males develop from unfertilized eggs (haploid) and females develop from fertilized eggs (diploid).
- Plants:
- Plants have diverse sex determination mechanisms. Some have genetic sex determination with sex chromosomes, while others have environmental sex determination, where environmental factors such as temperature, light, or chemical signals determine sex. Some plants can also change sex in response to environmental conditions.
These examples highlight the diversity of sex determination mechanisms in nature, reflecting the complex interplay between genetics, development, and the environment in determining an organism’s sex.
Sex-linked, sex-influenced, and sex-limited traits
Sex-linked, sex-influenced, and sex-limited traits are categories used to describe the inheritance patterns of certain traits based on an individual’s sex. These categories help explain how genes are expressed differently in males and females.
- Sex-Linked Traits:
- Definition: Sex-linked traits are traits that are located on the sex chromosomes (X and Y chromosomes in humans) and are inherited in a sex-dependent manner.
- Inheritance: Genes located on the X chromosome are considered X-linked. Since males have only one X chromosome (and one Y chromosome), they will express X-linked traits regardless of whether the allele is dominant or recessive. Females, with two X chromosomes, will express X-linked recessive traits only if they are homozygous for the recessive allele.
- Example: Color blindness and hemophilia are examples of X-linked recessive traits in humans.
- Sex-Influenced Traits:
- Definition: Sex-influenced traits are traits that are influenced by the presence of male or female sex hormones, resulting in different phenotypic expressions in males and females.
- Inheritance: The genes for sex-influenced traits are located on autosomal chromosomes, not on sex chromosomes. However, the expression of these genes can be influenced by sex hormones.
- Example: Male pattern baldness is a sex-influenced trait in humans. The gene for baldness is located on an autosome, but its expression is influenced by the presence of male sex hormones (androgens), leading to a higher incidence in males.
- Sex-Limited Traits:
- Definition: Sex-limited traits are traits that are expressed in only one sex, even though the genes for these traits may be present in both sexes.
- Inheritance: The genes for sex-limited traits are typically located on autosomal chromosomes, and their expression is controlled by sex-specific factors.
- Example: Milk production in mammals is a sex-limited trait. While both male and female mammals have the genes for producing milk, only females (usually after giving birth) are able to express this trait.
These categories help illustrate the complex interactions between genetics, hormones, and development in determining the expression of traits in males and females.
Dosage compensation and Y-linked characteristics
Dosage compensation is the process by which organisms equalize the expression of genes between individuals with different numbers of sex chromosomes. This is necessary because females typically have two X chromosomes (XX) and males have one X and one Y chromosome (XY) in many species, including humans. If dosage compensation did not occur, females would produce twice as much of the proteins encoded by X-linked genes as males, which could lead to imbalances in gene expression.
In mammals, including humans, dosage compensation is achieved by inactivating one of the X chromosomes in females. This process, known as X chromosome inactivation or lyonization, ensures that both males and females have only one active X chromosome in each cell. The inactivated X chromosome forms a dense structure called a Barr body, which is transcriptionally inactive.
Y-linked characteristics, on the other hand, are traits that are determined by genes located on the Y chromosome. Because the Y chromosome is much smaller than the X chromosome and contains fewer genes, Y-linked characteristics are relatively rare compared to X-linked characteristics. Y-linked traits are passed down from fathers to sons and are not present in females.
Examples of Y-linked characteristics in humans include male-pattern baldness and the presence of the SRY gene, which is responsible for triggering male development. Since females do not have a Y chromosome, they do not inherit Y-linked characteristics.
Quantitative Genetics
Quantitative traits and polygenic inheritance
Quantitative traits are traits that show continuous variation, meaning they can be measured along a continuum and are influenced by multiple genes and environmental factors. Examples of quantitative traits include height, weight, blood pressure, and IQ. These traits are often controlled by the combined effects of many genes, rather than being determined by a single gene with large effects.
Polygenic inheritance refers to the inheritance of quantitative traits that are controlled by multiple genes. Each gene contributes a small amount to the overall phenotype, and the effects of these genes are additive. The more genes that contribute to a trait, the more continuous the distribution of the trait will be in a population.
The concept of polygenic inheritance can be illustrated with the example of height in humans. Height is a quantitative trait influenced by many genes, each with a small effect. These genes may code for proteins involved in bone growth, hormone regulation, and other biological processes relevant to growth. The combined effects of these genes, along with environmental factors such as nutrition and health, determine an individual’s height.
Polygenic traits often exhibit a bell-shaped distribution when plotted in a population, known as a normal distribution or Gaussian distribution. The distribution reflects the combined effects of multiple genes and environmental factors on the trait. Polygenic inheritance contributes to the continuous variation seen in many traits and is important in understanding complex traits and diseases in populations.
Statistical methods to analyze quantitative characteristics
Statistical methods are essential for analyzing quantitative characteristics, which are traits that can be measured and vary continuously. Here are some common statistical methods used to analyze quantitative characteristics:
- Descriptive Statistics: Descriptive statistics are used to summarize and describe the main features of a dataset. Measures such as mean, median, mode, range, variance, and standard deviation provide information about the central tendency, dispersion, and shape of the data distribution.
- Graphical Representation: Graphical methods, such as histograms, box plots, and scatter plots, are used to visualize the distribution of quantitative data and identify patterns, outliers, and relationships between variables.
- Inferential Statistics: Inferential statistics are used to make inferences or predictions about a population based on a sample of data. Common inferential statistical tests for analyzing quantitative characteristics include:
- T-tests: Used to compare the means of two groups.
- Analysis of Variance (ANOVA): Used to compare the means of three or more groups.
- Regression Analysis: Used to model the relationship between a dependent variable and one or more independent variables.
- Correlation Analysis: Used to measure the strength and direction of the relationship between two quantitative variables.
- Chi-Square Test: Used to test the independence of two categorical variables.
- Linear Models: Linear regression is a common method used to model the relationship between a dependent variable and one or more independent variables. It assumes a linear relationship between the variables and is often used for prediction and hypothesis testing.
- Non-Parametric Methods: Non-parametric methods, such as the Mann-Whitney U test and the Kruskal-Wallis test, are used when data do not meet the assumptions of parametric tests (e.g., normality, homogeneity of variance).
- Multivariate Analysis: Multivariate analysis techniques, such as principal component analysis (PCA) and factor analysis, are used to analyze relationships among multiple variables and identify underlying patterns in the data.
- Survival Analysis: Survival analysis is used to analyze time-to-event data, such as time until death or time until a disease recurrence, and is often used in medical research and epidemiology.
These statistical methods provide researchers with tools to analyze, interpret, and draw conclusions from quantitative data, helping to understand the characteristics and relationships within a dataset.
Population Genetics
Allelic frequency and Hardy-Weinberg Law
Allelic frequency refers to the proportion of a particular allele in a population’s gene pool. It is calculated by dividing the number of copies of a specific allele by the total number of alleles at that genetic locus in the population. Allelic frequencies are important in population genetics as they help to understand genetic diversity, evolutionary processes, and the inheritance of genetic traits.
The Hardy-Weinberg law, also known as the Hardy-Weinberg equilibrium (HWE), describes the relationship between allelic frequencies and genotypic frequencies in a population that is not evolving. It states that under certain conditions, the frequencies of alleles and genotypes in a population will remain constant from generation to generation.
The Hardy-Weinberg law is based on several assumptions:
- Random mating: Individuals in the population mate randomly with respect to the gene in question.
- No mutation: There is no change in allele frequencies due to new mutations.
- No migration: There is no migration into or out of the population.
- Large population size: The population is large enough to prevent random genetic drift from significantly altering allele frequencies.
- No natural selection: There is no differential survival or reproductive success based on genotype.

This equation demonstrates that in a population at Hardy-Weinberg equilibrium, the frequencies of the three genotypes will stabilize and remain constant over time, as long as the assumptions of the Hardy-Weinberg equilibrium are met. Deviations from Hardy-Weinberg equilibrium can indicate evolutionary processes such as natural selection, genetic drift, mutation, or migration.
Applications of Hardy-Weinberg Law
The Hardy-Weinberg law has several important applications in population genetics and evolutionary biology:
- Estimating Allelic Frequencies: The Hardy-Weinberg law can be used to estimate allelic frequencies in a population based on genotype frequencies. By rearranging the equation one can solve for � and � to determine the frequencies of the two alleles.
- Testing for Evolutionary Forces: Deviations from Hardy-Weinberg equilibrium can indicate the presence of evolutionary forces such as natural selection, genetic drift, mutation, or migration. For example, an excess of homozygotes (observed �2 or �2 is significantly higher than expected) could suggest inbreeding or natural selection against heterozygotes.
- Estimating the Frequency of Carriers: In the case of genetic diseases caused by recessive alleles, the Hardy-Weinberg law can be used to estimate the frequency of carriers (heterozygotes) in a population. This information is important for genetic counseling and disease prevention programs.
- Understanding Genetic Variation: The Hardy-Weinberg law helps us understand the role of genetic variation in populations. It shows that even if a population is initially in Hardy-Weinberg equilibrium, evolutionary forces can cause allele frequencies to change over time.
- Detecting Non-Random Mating: Hardy-Weinberg equilibrium assumes random mating. Deviations from equilibrium can indicate non-random mating patterns, such as assortative mating (individuals preferentially mate with similar individuals) or disassortative mating (individuals preferentially mate with dissimilar individuals).
- Conservation Biology: The Hardy-Weinberg law is used in conservation biology to assess the genetic health of endangered populations. Deviations from Hardy-Weinberg equilibrium can indicate low genetic diversity, which may threaten the long-term survival of a population.
Overall, the Hardy-Weinberg law provides a valuable framework for understanding the dynamics of allele frequencies in populations and the forces that drive evolutionary change.
Natural selection, mutation, genetic drift, and migration
Natural selection, mutation, genetic drift, and migration are four key evolutionary forces that can lead to changes in allele frequencies in populations over time:
- Natural Selection:
- Definition: Natural selection is the process by which organisms with certain heritable traits are more likely to survive and reproduce than organisms with other traits, leading to changes in the frequency of those traits in a population over generations.
- Mechanism: Natural selection acts on phenotypic variation within a population. Traits that increase an organism’s fitness (ability to survive and reproduce) are more likely to be passed on to the next generation, leading to an increase in the frequency of those traits in the population.
- Types: Natural selection can be classified into three main types: directional selection (favoring one extreme phenotype), stabilizing selection (favoring intermediate phenotypes), and disruptive selection (favoring both extreme phenotypes).
- Mutation:
- Definition: Mutation is a change in the DNA sequence of an organism. Mutations can create new alleles, leading to genetic variation within a population.
- Effect on Allele Frequencies: Mutations introduce new alleles into a population. While most mutations are neutral or deleterious, some mutations can be beneficial and increase in frequency through natural selection.
- Genetic Drift:
- Definition: Genetic drift is the random change in allele frequencies in a population due to chance events. Genetic drift is most pronounced in small populations.
- Effect on Allele Frequencies: Genetic drift can lead to the loss of alleles in a population, especially in small populations where chance events can have a significant impact. Genetic drift can also lead to the fixation of alleles, where one allele becomes the only allele at a particular locus in the population.
- Migration (Gene Flow):
- Definition: Migration, also known as gene flow, is the movement of alleles between populations. Migration can introduce new alleles into a population or alter the frequencies of existing alleles.
- Effect on Allele Frequencies: Migration can increase genetic variation within a population by introducing new alleles. It can also reduce genetic differences between populations, leading to genetic homogenization.
These evolutionary forces interact with each other and with environmental factors to shape the genetic diversity and adaptation of populations over time. Understanding these forces is essential for studying the mechanisms of evolution and how populations respond to environmental changes.
Basic Principles of Heredity
Mendelian genetics: experiments and terminology
Mendelian genetics, based on the work of Gregor Mendel in the 19th century, describes the inheritance of traits controlled by a single gene with two alleles, one dominant and one recessive. Mendel’s experiments with pea plants laid the foundation for modern genetics and introduced key concepts and terminology:
- Mendel’s Experiments:
- Mendel conducted hybridization experiments with pea plants, studying seven different traits including seed color, seed shape, flower color, flower position, pod shape, pod color, and stem height.
- He crossed plants with contrasting traits (e.g., yellow seeds with green seeds) and studied the inheritance patterns in the offspring (F1 generation) and subsequent generations (F2 generation).
- Key Concepts:
- Dominant and Recessive Traits: Mendel observed that some traits (e.g., yellow seed color) were dominant and masked the expression of other traits (e.g., green seed color, recessive).
- Segregation: Mendel proposed the Law of Segregation, which states that each individual has two alleles for each gene, which segregate during gamete formation, so each gamete carries only one allele for each gene.
- Independent Assortment: Mendel proposed the Law of Independent Assortment, which states that genes for different traits segregate independently of each other during gamete formation, leading to the random assortment of alleles into gametes.
- Terminology:
- Allele: Different forms of a gene that can occupy the same locus on homologous chromosomes (e.g., yellow and green alleles for seed color).
- Genotype: The genetic makeup of an individual, typically represented by letters (e.g., YY, Yy, yy for seed color).
- Phenotype: The observable traits of an individual, determined by its genotype and environmental factors (e.g., yellow or green seed color).
- Homozygous: When an individual has two identical alleles for a particular gene (e.g., YY or yy).
- Heterozygous: When an individual has two different alleles for a particular gene (e.g., Yy).
Mendel’s experiments and the principles of Mendelian genetics laid the groundwork for our understanding of inheritance patterns and provided a framework for studying more complex genetic traits and phenomena.
Mendel’s laws: monohybrid and dihybrid crosses
Mendel’s laws, based on his experiments with pea plants, describe the principles of inheritance for single gene traits (monohybrid crosses) and two gene traits (dihybrid crosses). These laws are fundamental to the study of genetics. Here’s a brief overview of Mendel’s laws and how they apply to monohybrid and dihybrid crosses:
- Law of Segregation (First Law):
- Each individual has two alleles for each gene, one from each parent.
- These alleles segregate (separate) during gamete formation, so each gamete carries only one allele for each gene.
- This law explains the 3:1 ratio of dominant to recessive phenotypes in the F2 generation of a monohybrid cross.
- Law of Independent Assortment (Second Law):
- Genes for different traits are inherited independently of each other.
- This law explains the 9:3:3:1 ratio of phenotypes in the F2 generation of a dihybrid cross involving two different traits.
Monohybrid Cross:
- A monohybrid cross involves the inheritance of a single gene with two alleles (e.g., A and a).
- The Punnett square is a commonly used tool to predict the outcomes of monohybrid crosses.
- For example, in a monohybrid cross between two heterozygous (Aa) individuals, the offspring would be expected to have a genotype ratio of 1:2:1 (AA:Aa:aa) and a phenotype ratio of 3:1 (dominant:recessive).
Dihybrid Cross:
- A dihybrid cross involves the inheritance of two genes, each with two alleles (e.g., A/a and B/b).
- The Law of Independent Assortment predicts that the alleles for each gene segregate independently of alleles for other genes.
- The Punnett square can also be used to predict the outcomes of dihybrid crosses, considering the possible combinations of alleles from each parent.
- For example, in a dihybrid cross between two heterozygous (AaBb) individuals, the offspring would be expected to have a genotype ratio of 1:2:1:2:4:2:1:2:1 (AABB:AABb:AAbb:AaBB:AaBb:Aabb:aaBB:aaBb:aabb) and a phenotype ratio of 9:3:3:1 (both dominant:one dominant, one recessive:one dominant, both recessive:both recessive).
Deviations from Mendel’s ratios
While Mendel’s laws provide a useful framework for understanding inheritance patterns, deviations from Mendel’s ratios can occur due to various factors. Some common reasons for deviations from Mendel’s ratios include:
- Incomplete Dominance: In incomplete dominance, the heterozygote phenotype is intermediate between the two homozygotes. For example, in snapdragons, the flower color is not purely red (RR) or white (rr) but pink (Rr).
- Codominance: In codominance, both alleles are fully expressed in the heterozygote, resulting in a phenotype that shows aspects of both alleles. An example is blood type in humans, where the AB blood type has both A and B antigens.
- Multiple Alleles: Some traits are controlled by multiple alleles, where more than two alleles exist for a particular gene in a population. However, an individual can still only have two alleles. ABO blood group is an example of multiple alleles.
- Pleiotropy: Some genes have multiple effects on an organism’s phenotype. For example, a single gene mutation can affect multiple traits, leading to unexpected phenotypic ratios.
- Epistasis: Epistasis occurs when the expression of one gene is influenced by another gene. This can lead to modified phenotypic ratios, as the presence of one allele masks the effects of alleles at another locus.
- Environmental Influences: Environmental factors can also influence phenotypic expression. For example, temperature can affect the coloration of Siamese cats and Himalayan rabbits.
- Linked Genes: Genes located close to each other on the same chromosome tend to be inherited together (linkage). This can lead to deviations from expected Mendelian ratios if two genes are closely linked and do not assort independently.
- Gene Interactions: Interaction between genes at different loci (epistasis) or within the same locus (complementation, redundancy) can affect phenotypic ratios.
These deviations highlight the complexity of genetic inheritance and the need to consider various factors beyond simple Mendelian ratios when studying inheritance patterns.
Genetic interactions
Genetic interactions refer to the ways in which different genes or alleles interact with each other to produce a particular phenotype. Here are explanations of epistasis, pleiotropy, penetrance, and expressivity:
- Epistasis:
- Definition: Epistasis is a genetic interaction in which the effects of one gene mask or modify the effects of another gene at a different locus.
- Types of Epistasis: There are several types of epistasis, including dominant epistasis (when the presence of one allele masks the effects of other alleles at different loci), recessive epistasis (when the presence of two recessive alleles at one locus masks the effects of alleles at another locus), and duplicate recessive epistasis (when the presence of either of two recessive alleles at one locus masks the effects of alleles at another locus).
- Pleiotropy:
- Definition: Pleiotropy occurs when a single gene influences multiple, seemingly unrelated phenotypic traits.
- Example: A mutation in the gene responsible for sickle cell anemia not only affects red blood cell shape but also confers resistance to malaria.
- Penetrance:
- Definition: Penetrance refers to the proportion of individuals with a specific genotype who express the expected phenotype. Incomplete penetrance occurs when individuals with the genotype do not express the phenotype.
- Example: In some cases of hereditary breast cancer, individuals with a mutation in the BRCA1 gene may or may not develop breast cancer, indicating incomplete penetrance.
- Expressivity:
- Definition: Expressivity refers to the degree or extent to which a genotype is expressed in an individual. It can vary from mild to severe, even among individuals with the same genotype.
- Example: Neurofibromatosis type 1 (NF1) is characterized by tumors along nerves. The expressivity of NF1 can vary widely, with some individuals having only a few tumors while others have many.
Understanding genetic interactions is important for elucidating the complexity of inheritance patterns and how genes interact to produce phenotypic diversity. These concepts help explain why individuals with the same genotype can exhibit different phenotypes and contribute to our understanding of genetic diseases and traits.
Multiple alleles
Multiple alleles refer to the existence of more than two alleles (variants of a gene) at a specific locus (location on a chromosome) in a population. However, each individual organism still carries only two alleles, one inherited from each parent. The concept of multiple alleles is important in understanding the diversity of traits within populations and the inheritance patterns of certain genetic traits.
Examples of traits controlled by multiple alleles include the ABO blood group system in humans and coat color in rabbits. In the ABO blood group system, there are three alleles (IA, IB, i) that determine the A, B, and O blood types. The IA and IB alleles are codominant, while the i allele is recessive. This results in four possible blood types (A, B, AB, O) based on the combinations of these three alleles.
Multiple alleles can result in complex inheritance patterns. For example, in the ABO blood group system, the IA and IB alleles are codominant, meaning that individuals with the IAIB genotype will have the AB blood type, which expresses both A and B antigens. This is different from traits with only two alleles, where the heterozygous genotype typically exhibits an intermediate phenotype.
Understanding multiple alleles is important in genetics and evolutionary biology, as they contribute to the genetic diversity of populations and can influence the prevalence of certain traits in a population.