
Identifying Paralogs and Orthologs via COGs and KOGs databases
October 8, 2007Orthologs and Paralogs defined as
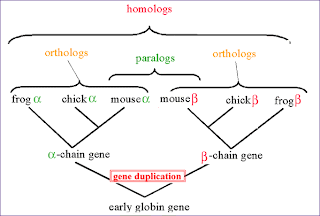
Orthologs: similar sequences or genes in different species that arose through speciation and mutation and not from gene duplication.
Paralogs: Related genes(or proteins) in the same genome. The related genes have arisen by gene duplication.
COG and KOG databases:
The COG(Clusters of Orthologous Groups) and KOG (euKaryotic Orthologous Groups) databases have been constructed using a careful analysis of BLAST hits.
First, low-complexity sequence regions and commonly occuring domains are masked to prevent spurious hits and also to improve the the statistical score analysis (E-values).
All gene sequences from one genome are then scanned against all from another genome, noting the best-scoring BLAST hits for each gene, and this is repeated for all possible pairs.
Paralogous genes within a genome that result from gene duplication since divergence of two species are identified as those that give a better-scoring BLAST hit with each other than their BLAST hits with the other genome.
Orthologus genes are found as groups of genes from different genomes that are reciprocal BLAST hits of each other.
All sequences in a COG or a KOG are assumed to have a related function, and thus the method can be used to predict gene and protein function.
