
Introduction to Biology for Bioinformatics
February 29, 2024 Off By adminCourse Description:
This course provides computer science students with a foundational understanding of biology necessary for bioinformatics. Students will learn key biological concepts and how they relate to computational methods used in analyzing biological data. Topics include molecular biology, genetics, cellular biology, and bioinformatics tools and techniques. This course will provide computer science students with a solid foundation in biology, preparing them for more advanced courses in bioinformatics.
Prerequisites:
- Basic programming skills (e.g., proficiency in Python)
- Familiarity with data structures and algorithms
- Understanding of mathematical concepts (e.g., statistics, linear algebra)
Table of Contents
Introduction to Bioinformatics
Overview of bioinformatics and its applications
Bioinformatics is an interdisciplinary field that combines biology, computer science, mathematics, and statistics to analyze and interpret biological data. It involves the development and application of computational tools and techniques to understand biological processes. Here’s an overview of bioinformatics and its applications:
- Sequence Analysis: One of the fundamental areas of bioinformatics is the analysis of biological sequences, such as DNA, RNA, and proteins. This includes sequence alignment, similarity searching, and prediction of protein structures and functions.
- Genome Annotation: Bioinformatics plays a crucial role in annotating genomes by identifying genes, regulatory elements, and other functional elements within DNA sequences.
- Phylogenetics: Bioinformatics is used to reconstruct evolutionary relationships among organisms based on genetic data, such as DNA sequences, to understand evolutionary processes and classify species.
- Structural Biology: Bioinformatics tools are used to predict and analyze the three-dimensional structures of proteins and nucleic acids, which is crucial for understanding their functions and interactions.
- Systems Biology: Bioinformatics is applied to study biological systems as a whole, including modeling complex biological processes, such as metabolic pathways and regulatory networks.
- Functional Genomics: Bioinformatics helps in understanding the functions of genes and proteins on a genome-wide scale, including gene expression analysis, regulatory element identification, and protein interaction networks.
- Metagenomics: Bioinformatics is used to analyze complex microbial communities by sequencing and analyzing the genetic material directly from environmental samples, providing insights into microbial diversity and function.
- Drug Discovery and Development: Bioinformatics plays a key role in drug discovery by predicting drug-target interactions, virtual screening of compounds, and understanding the mechanisms of drug resistance.
- Personalized Medicine: Bioinformatics is used to analyze individual genetic variations that influence disease susceptibility, drug response, and other personalized healthcare aspects.
- Clinical Genomics: Bioinformatics is applied in clinical settings for diagnosing genetic diseases, identifying disease markers, and predicting patient outcomes based on genetic information.
Overall, bioinformatics has revolutionized biological research by providing powerful tools and methods for analyzing and interpreting biological data, leading to a deeper understanding of living systems and their applications in various fields, including healthcare, agriculture, and environmental science.

bioinformatics application
Importance of biology in bioinformatics
Biology is foundational to bioinformatics, as bioinformatics is the application of computational tools and techniques to understand biological data. Here are some key reasons why biology is crucial in bioinformatics:
- Understanding Biological Context: Biology provides the context and background knowledge necessary to interpret and analyze biological data. Understanding biological processes, such as gene regulation, protein structure, and cellular pathways, is essential for meaningful analysis.
- Data Interpretation: Biological knowledge helps in interpreting the results of bioinformatics analyses. For example, knowing the function of a gene or protein can help interpret its role in a biological process or disease.
- Tool Development: Biological insights drive the development of bioinformatics tools and algorithms. Understanding biological processes helps in designing tools that are relevant and effective for analyzing biological data.
- Hypothesis Generation: Biology provides the basis for hypothesis generation in bioinformatics research. By understanding biological systems, researchers can formulate hypotheses about the functions and interactions of genes, proteins, and other biological molecules.
- Validation of Results: Biological knowledge is essential for validating the results of bioinformatics analyses. Experimental validation is often necessary to confirm the findings of computational analyses, and biological knowledge guides the design and interpretation of these experiments.
- Integration of Data: Biology provides the framework for integrating data from different sources and levels of biological organization. This integration is essential for understanding complex biological systems.
- Biological Data Management: Understanding biological data requires knowledge of the types of data generated in biological experiments, such as sequencing data, microarray data, and protein structures. This knowledge is essential for managing and analyzing biological data effectively.
In summary, biology is essential in bioinformatics because it provides the context, interpretation, and foundation for the analysis and understanding of biological data using computational tools and techniques.
Basics of Molecular Biology
Introduction to DNA, RNA, and proteins
DNA, RNA, and proteins are fundamental molecules in biology, each playing crucial roles in the structure, function, and regulation of living organisms. Here’s a brief introduction to each:
- DNA (Deoxyribonucleic Acid):
- DNA is a double-stranded molecule that carries the genetic instructions used in the growth, development, functioning, and reproduction of all known living organisms and many viruses.
- It consists of two long chains of nucleotides twisted into a double helix and held together by hydrogen bonds between complementary base pairs (adenine with thymine, and cytosine with guanine).
- DNA is found in the nucleus of eukaryotic cells and in the cytoplasm of prokaryotic cells. It contains the genes that encode proteins and non-coding RNAs, which play essential roles in cellular processes.
- RNA (Ribonucleic Acid):
- RNA is a single-stranded molecule that is involved in various biological roles, including coding, decoding, regulation, and expression of genes.
- It is made up of nucleotides, similar to DNA, but with uracil (U) replacing thymine (T) as one of the bases. The other bases are adenine, cytosine, and guanine.
- RNA is produced from DNA through a process called transcription and is involved in translating the genetic code into proteins (messenger RNA or mRNA), regulating gene expression (microRNA or miRNA), and catalyzing biochemical reactions (ribosomal RNA or rRNA and transfer RNA or tRNA).
- Proteins:
- Proteins are large, complex molecules that are essential for the structure, function, and regulation of the body’s tissues and organs.
- They are made up of amino acids, which are linked together in a specific sequence determined by the genetic code.
- Proteins perform a wide variety of functions in the body, including catalyzing biochemical reactions, providing structural support to cells and tissues, transporting molecules, and serving as signaling molecules.
In summary, DNA stores genetic information, RNA helps in the expression and regulation of genes, and proteins carry out the functions specified by the genetic information. Together, these molecules form the basis of the central dogma of molecular biology, which describes the flow of genetic information from DNA to RNA to protein.
Central dogma of molecular biology
The central dogma of molecular biology is a fundamental concept that describes the flow of genetic information in biological systems. It was first proposed by Francis Crick in 1958 and later expanded upon. The central dogma states that genetic information flows from DNA to RNA to proteins, and it serves as the basis for understanding how genetic information is stored, replicated, and expressed in living organisms.
Here’s a brief overview of each step in the central dogma:
- DNA (Deoxyribonucleic Acid): DNA is a molecule that contains the genetic instructions for the development, functioning, growth, and reproduction of all known living organisms. It is composed of two strands that are arranged in a double helix structure. The sequence of nucleotides (adenine, thymine, cytosine, and guanine) in DNA determines the genetic information that an organism inherits.
- Transcription: The first step in the central dogma is transcription, during which a segment of DNA is copied into a complementary RNA molecule. This process is catalyzed by an enzyme called RNA polymerase and involves the formation of a single-stranded RNA molecule that is complementary to one of the DNA strands. The RNA molecule produced is called messenger RNA (mRNA).
- RNA (Ribonucleic Acid): RNA is a molecule that is similar to DNA but differs in several key ways. It is typically single-stranded and contains the nucleotides adenine, uracil, cytosine, and guanine. RNA plays a crucial role in protein synthesis and other cellular processes.
- Translation: The final step in the central dogma is translation, during which the genetic information encoded in mRNA is used to synthesize a protein. This process occurs on ribosomes, which are cellular structures composed of RNA and proteins. Transfer RNA (tRNA) molecules carry specific amino acids to the ribosome, where they are linked together to form a protein according to the sequence of codons (three-nucleotide sequences) in the mRNA.
In summary, the central dogma of molecular biology describes the flow of genetic information from DNA to RNA to proteins, outlining the processes of transcription and translation that are essential for gene expression and protein synthesis in living organisms.

Genetic code and gene expression
The genetic code is the set of rules by which information encoded in genetic material (DNA or RNA sequences) is translated into proteins (sequences of amino acids). It is the basis for gene expression, which is the process by which information from a gene is used to synthesize a functional gene product, such as a protein.
The genetic code is composed of codons, which are sequences of three nucleotides (triplets) that specify a particular amino acid. There are 64 possible codons, but only 20 amino acids, so most amino acids are encoded by more than one codon (redundancy), except for methionine and tryptophan, which each have a single codon. Additionally, there are three stop codons (also called termination codons or nonsense codons) that signal the end of protein synthesis.
Gene expression involves two main processes: transcription and translation.
- Transcription: In transcription, the DNA sequence of a gene is copied into a complementary RNA molecule by an enzyme called RNA polymerase. This process occurs in the nucleus of eukaryotic cells (or in the cytoplasm of prokaryotic cells) and produces a molecule called messenger RNA (mRNA). The mRNA carries the genetic information from the DNA to the ribosome for protein synthesis.
- Translation: In translation, the mRNA is read by ribosomes in the cytoplasm, and the genetic code is used to synthesize a protein. Transfer RNA (tRNA) molecules carry specific amino acids to the ribosome, and each tRNA molecule recognizes a specific codon on the mRNA through complementary base pairing. As the ribosome moves along the mRNA, it joins amino acids together in the order specified by the mRNA codons, forming a polypeptide chain that will fold into a functional protein.
Overall, the genetic code and gene expression are essential processes that enable cells to produce the proteins necessary for their structure, function, and regulation. They are fundamental to understanding how genetic information is used to create the diversity of life.
Genetics and Genomics
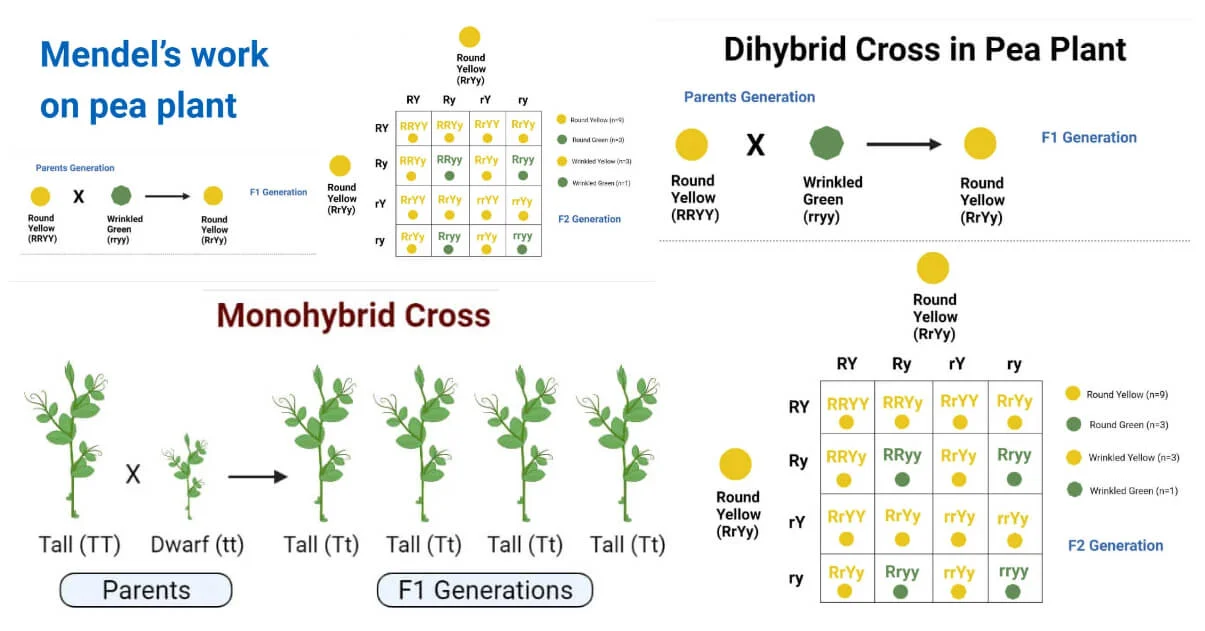
Mendelian genetics and inheritance patterns
Mendelian genetics, named after the pioneering work of Gregor Mendel in the 19th century, describes the inheritance patterns of certain traits from parents to offspring. Mendel’s experiments with pea plants led to the discovery of basic principles of inheritance, which laid the foundation for modern genetics.
Here are some key concepts of Mendelian genetics and inheritance patterns:
- Dominant and Recessive Alleles: Mendel observed that traits are controlled by discrete units of inheritance, which we now call genes. Each gene exists in two alternative forms called alleles. For example, in the case of flower color in pea plants, there are two alleles: one for purple flowers (dominant allele, usually denoted by a capital letter, such as “P”) and one for white flowers (recessive allele, usually denoted by a lowercase letter, such as “p”). The dominant allele masks the expression of the recessive allele in heterozygous individuals.
- Genotype and Phenotype: An organism’s genotype refers to the specific alleles it carries for a particular trait, while its phenotype refers to the physical expression of that trait. For example, an individual with the genotype “PP” or “Pp” will have purple flowers (phenotype), while an individual with the genotype “pp” will have white flowers.
- Punnett Squares: Punnett squares are diagrams used to predict the possible genotypes and phenotypes of offspring from a genetic cross between two individuals. They are based on the principles of probability and help illustrate Mendelian inheritance patterns.
- Genetic Crosses: Genetic crosses involve mating individuals with known genotypes to study the inheritance of specific traits. For example, a cross between two heterozygous purple-flowered pea plants (Pp x Pp) would produce offspring with a 3:1 ratio of purple to white flowers, following Mendel’s principle of segregation.
- Principle of Segregation: According to Mendel’s principle of segregation, the two alleles for a gene segregate (separate) during the formation of gametes (sperm and egg cells), so that each gamete carries only one allele for each gene. This explains why offspring inherit one allele from each parent.
- Principle of Independent Assortment: The principle of independent assortment states that genes located on different chromosomes assort independently during the formation of gametes. This means that the inheritance of one gene does not influence the inheritance of another gene, leading to the formation of different combinations of alleles in offspring.
Mendelian genetics provides a basic understanding of inheritance patterns, but it is important to note that many traits are influenced by multiple genes and environmental factors, leading to more complex inheritance patterns than those described by Mendel.
Introduction to genomics and sequencing technologies
Genomics is the study of an organism’s entire genome, which includes all of its genes and non-coding sequences of DNA. Genomics aims to understand the structure, function, and evolution of genomes, as well as how genes interact to produce traits and how genetic variation contributes to disease and other phenotypes.
Sequencing technologies play a crucial role in genomics by enabling the determination of the order of nucleotides (A, T, C, G) in an organism’s DNA. Here is an overview of genomics and sequencing technologies:
- DNA Sequencing: DNA sequencing is the process of determining the precise order of nucleotides in a DNA molecule. The first methods developed for DNA sequencing were labor-intensive and time-consuming, but advancements in technology have led to the development of high-throughput sequencing techniques that can sequence entire genomes rapidly and cost-effectively.
- Next-Generation Sequencing (NGS): NGS technologies, also known as high-throughput sequencing technologies, have revolutionized genomics by enabling the rapid sequencing of large amounts of DNA. NGS methods, such as Illumina sequencing, use parallel sequencing of millions of DNA fragments to generate massive amounts of sequencing data quickly.
- Third-Generation Sequencing: Third-generation sequencing technologies, such as PacBio and Oxford Nanopore sequencing, offer long-read sequencing capabilities, which can provide a more complete picture of complex genomes, including structural variations and repetitive regions that are challenging for short-read sequencing methods.
- Applications of Genomics: Genomics has applications in various fields, including medicine, agriculture, evolutionary biology, and environmental science. In medicine, genomics is used for personalized medicine, genetic disease screening, and understanding the genetic basis of diseases such as cancer.
- Bioinformatics in Genomics: Bioinformatics plays a crucial role in genomics by providing tools and algorithms for analyzing and interpreting genomic data. Bioinformaticians use computational methods to assemble and annotate genomes, identify genetic variants, and analyze gene expression and regulation.
- Future Directions: The field of genomics continues to advance rapidly, with ongoing efforts to sequence and analyze the genomes of diverse organisms, including humans, plants, animals, and microorganisms. Emerging technologies and computational methods are expanding our understanding of genomic complexity and its implications for biology and medicine.
In summary, genomics and sequencing technologies have revolutionized our understanding of the genetic basis of life and are driving advances in biology, medicine, and other fields. Continued advancements in sequencing technologies and bioinformatics are expected to further enhance our understanding of genomes and their functions.

Evolution-of-sequencing technology
Genome organization and variation
Genome organization refers to the arrangement of DNA within an organism’s genome. The genome of an organism is organized into chromosomes, which are long strands of DNA that contain many genes. Genome organization can vary among different organisms, but there are some common features:
- Chromosomes: In most organisms, the genome is organized into chromosomes, which are structures that contain the genetic material. Chromosomes can vary in number and size among different species.
- Genes: Genes are the units of heredity that contain the instructions for making proteins. Genes are located on chromosomes and are organized into regions called exons (coding regions) and introns (non-coding regions).
- Non-coding DNA: In addition to genes, the genome contains non-coding DNA sequences that do not code for proteins. These sequences can have regulatory functions, such as controlling when and where genes are expressed.
- Repetitive DNA: The genome contains repetitive DNA sequences that are repeated many times. These sequences can be interspersed throughout the genome or clustered in specific regions.
- Structural Variation: Structural variation refers to differences in the structure of the genome, such as insertions, deletions, duplications, and inversions of DNA segments. These variations can affect gene expression and contribute to genetic diversity among individuals.
- Single Nucleotide Polymorphisms (SNPs): SNPs are variations in a single nucleotide that occur at specific positions in the genome. SNPs are the most common type of genetic variation in the human genome and can be used as genetic markers in studies of genetic diversity and disease susceptibility.
- Copy Number Variation (CNV): CNV refers to variations in the number of copies of a particular DNA segment that are present in the genome. CNVs can range in size from kilobases to megabases and can influence gene expression and phenotype.
Genome variation refers to differences in the DNA sequence between individuals of the same species. Genome variation can arise from mutations, which are changes in the DNA sequence that can be inherited or occur spontaneously. Genome variation is important for evolution and adaptation, as it can lead to differences in traits among individuals and populations.
Studying genome organization and variation is important for understanding the genetic basis of traits and diseases, as well as for evolutionary and population genetics research. Advances in sequencing technologies and computational methods have made it possible to study genome organization and variation in unprecedented detail, leading to new insights into the genetic diversity of life.
Cellular Biology
Cell structure and function
Cell structure and function are fundamental concepts in biology, as cells are the basic units of life and carry out all the functions necessary for an organism to survive. Here is an overview of cell structure and function:
- Cell Membrane: The cell membrane, also known as the plasma membrane, is a lipid bilayer that surrounds the cell and separates it from its environment. It regulates the passage of substances into and out of the cell and plays a role in cell communication and signaling.
- Cytoplasm: The cytoplasm is the gel-like substance inside the cell that contains organelles, cytoskeleton, and various molecules. It is where many cellular processes, such as metabolism and protein synthesis, occur.
- Nucleus: The nucleus is a membrane-bound organelle that contains the cell’s genetic material, DNA. It controls the cell’s activities by regulating gene expression and is essential for cell division.
- Organelles: Organelles are specialized structures within the cell that perform specific functions. Examples include mitochondria (energy production), endoplasmic reticulum (protein synthesis and lipid metabolism), Golgi apparatus (protein processing and packaging), and lysosomes (digestion of cellular waste).
- Cytoskeleton: The cytoskeleton is a network of protein filaments that provides structural support to the cell and helps maintain its shape. It is also involved in cell movement and intracellular transport.
- Mitochondria: Mitochondria are organelles that are responsible for generating energy in the form of adenosine triphosphate (ATP) through the process of cellular respiration. They are often referred to as the “powerhouses” of the cell.
- Endoplasmic Reticulum: The endoplasmic reticulum (ER) is a network of membranes that is involved in protein synthesis, lipid metabolism, and the transport of proteins within the cell.
- Golgi Apparatus: The Golgi apparatus is a stack of membrane-bound vesicles that is involved in processing, packaging, and distributing proteins and lipids within the cell.
- Lysosomes: Lysosomes are membrane-bound organelles that contain enzymes responsible for breaking down cellular waste and foreign materials, such as bacteria.
- Cellular Processes: Cells carry out various processes to maintain homeostasis and respond to their environment, including metabolism (chemical reactions that provide energy and nutrients), growth and division (cell cycle), and communication with other cells (cell signaling).
Overall, the structure and function of cells are tightly regulated and coordinated to ensure the survival and proper functioning of organisms. Cells are highly specialized and can vary greatly in structure and function depending on their type and role in the body.

Animal-cell.
Cell cycle and cell division
The cell cycle is the series of events that take place in a cell leading to its division and duplication of its DNA to produce two daughter cells. It consists of several phases, including interphase (G1, S, and G2 phases) and mitotic phase (mitosis and cytokinesis). Here’s an overview of the cell cycle and cell division:
- Interphase:
- G1 Phase (Gap 1): The cell grows and carries out its normal functions. It prepares for DNA replication.
- S Phase (Synthesis): DNA replication occurs, resulting in the duplication of the genetic material.
- G2 Phase (Gap 2): The cell continues to grow and prepares for cell division.
- Mitotic Phase:
- Mitosis: The nucleus of the cell divides into two nuclei, each with the same number of chromosomes as the parent cell. Mitosis is divided into several stages: prophase, prometaphase, metaphase, anaphase, and telophase.
- Cytokinesis: The cytoplasm of the cell divides, resulting in two daughter cells.
After cytokinesis, the two daughter cells enter the G1 phase of the cell cycle, and the process repeats.
Cell division is essential for growth, development, and repair of multicellular organisms. It ensures that each new cell receives a complete set of genetic information and the necessary cellular components to function properly. Dysregulation of the cell cycle and cell division can lead to diseases such as cancer. Understanding the cell cycle and cell division is crucial for studying normal cellular processes and disease mechanisms.

Cell-Division
Introduction to microbiology and microbial genomics
Microbiology is the study of microorganisms, which are microscopic organisms such as bacteria, viruses, fungi, and protozoa. Microorganisms play critical roles in various biological processes, including nutrient cycling, symbiotic relationships, and disease. Microbiology encompasses several sub-disciplines, including medical microbiology, environmental microbiology, industrial microbiology, and microbial genetics.
Microbial genomics is a field of microbiology that focuses on the study of the genomes of microorganisms. It involves the sequencing, analysis, and comparison of microbial genomes to understand their genetic composition, evolution, and function. Microbial genomics has revolutionized our understanding of microbial diversity, evolution, and pathogenesis.
Key concepts in microbiology and microbial genomics include:
- Microbial Diversity: Microorganisms exhibit a vast diversity in terms of morphology, physiology, and genetic makeup. Microbial diversity is essential for ecosystem functioning and has implications for human health, agriculture, and industry.
- Microbial Ecology: Microbial ecology is the study of the interactions of microorganisms with their environment, including other organisms. It includes the study of microbial communities, such as the human microbiome and soil microbiome, and their roles in biogeochemical cycles.
- Microbial Pathogenesis: Some microorganisms are pathogens that can cause diseases in humans, animals, and plants. Microbial pathogenesis is the study of how pathogens cause disease and the host-pathogen interactions involved.
- Microbial Genetics: Microbial genetics focuses on the study of the genetic makeup of microorganisms, including their genomes, genetic variation, gene regulation, and genetic transfer mechanisms.
- Genome Sequencing and Analysis: Advances in sequencing technologies have enabled the sequencing of microbial genomes at a rapid pace and at a relatively low cost. Genome sequencing and analysis provide insights into the genetic basis of microbial traits, evolution, and adaptation.
- Biotechnological Applications: Microorganisms are used in various biotechnological applications, such as the production of antibiotics, enzymes, and biofuels. Microbial genomics plays a crucial role in understanding and optimizing these processes.
Overall, microbiology and microbial genomics are dynamic fields that continue to expand our understanding of the microbial world and its impact on diverse aspects of life on Earth.
Bioinformatics Tools and Databases
Introduction to bioinformatics databases (e.g., NCBI, Ensembl)
Bioinformatics databases are repositories of biological data that are organized, curated, and made available for researchers to access and analyze. These databases play a crucial role in biological research by providing a centralized and standardized source of information that can be used to study various aspects of genomics, proteomics, and other fields of biology. Here’s an introduction to some of the major bioinformatics databases:
- NCBI (National Center for Biotechnology Information): NCBI is a comprehensive resource for biological information, including nucleotide sequences (GenBank), protein sequences (RefSeq), and biomedical literature (PubMed). It also provides tools for sequence analysis, genome browsing, and data mining.
- Ensembl: Ensembl is a genome database that provides genome assemblies, gene annotations, and comparative genomics data for a wide range of species. It offers tools for visualizing and analyzing genomic data, such as the Ensembl Genome Browser.
- UniProt: UniProt is a comprehensive resource for protein sequence and functional information, providing access to protein sequences, annotations, and cross-references to other databases. It includes the Swiss-Prot and TrEMBL databases.
- GenBank: GenBank is a database of nucleotide sequences, maintained by the NCBI. It contains sequences submitted by researchers as well as those generated by large-scale sequencing projects, such as the Human Genome Project.
- PDB (Protein Data Bank): PDB is a database of 3D structural data for proteins and other biological macromolecules. It provides information on the structure, function, and interactions of proteins, as well as tools for visualizing and analyzing protein structures.
- KEGG (Kyoto Encyclopedia of Genes and Genomes): KEGG is a database of biological pathways and functional annotations for genomes. It provides information on metabolic pathways, regulatory networks, and other biological processes.
- Reactome: Reactome is a database of biological pathways and reactions, focusing on human biology. It provides detailed information on signaling pathways, metabolic pathways, and other cellular processes.
- EBI (European Bioinformatics Institute) Databases: EBI hosts several databases, including the European Nucleotide Archive (ENA), which stores nucleotide sequences, and ArrayExpress, which stores gene expression data. EBI also provides tools and resources for analyzing and visualizing biological data.
These databases, along with many others, provide valuable resources for researchers in bioinformatics, genomics, and related fields to access, analyze, and interpret biological data.

Basic bioinformatics tools for sequence analysis (e.g., BLAST, ClustalW)
Bioinformatics databases and tools are essential for storing, managing, and analyzing biological data, particularly DNA, RNA, and protein sequences. Here’s an introduction to some commonly used bioinformatics databases and tools:
- NCBI (National Center for Biotechnology Information): NCBI is a comprehensive resource for biological information, offering databases such as GenBank (a repository of DNA sequences), PubMed (a database of biomedical literature), and BLAST (Basic Local Alignment Search Tool for sequence similarity searches).
- Ensembl: Ensembl is a genome database and browser that provides access to annotated genome sequences for a wide range of species. It includes tools for exploring gene structures, variations, and comparative genomics.
- UniProt: UniProt is a comprehensive resource for protein sequence and annotation information, providing access to high-quality, curated protein sequences and functional information.
- GenBank: GenBank is a database of genetic sequences maintained by NCBI. It includes sequences submitted by researchers as well as sequences obtained from the scientific literature.
- BLAST (Basic Local Alignment Search Tool): BLAST is a widely used tool for comparing a query sequence against a database of known sequences to identify similar sequences. It is used for sequence similarity searches and can help identify homologous genes and proteins.
- ClustalW: ClustalW is a tool for multiple sequence alignment, which is used to align multiple sequences to identify regions of similarity and difference. It helps in studying evolutionary relationships and identifying conserved regions in sequences.
- ExPASy: ExPASy is a bioinformatics resource portal that provides access to a variety of tools and databases for protein analysis, including tools for sequence analysis, structure prediction, and post-translational modification prediction.
- KEGG (Kyoto Encyclopedia of Genes and Genomes): KEGG is a database of biological pathways and networks, providing information on the functional and metabolic pathways of genes and molecules in various organisms.
These databases and tools are essential for bioinformatics research, enabling researchers to access, analyze, and interpret biological data to gain insights into the structure, function, and evolution of genes, proteins, and genomes.
Introduction to genome browsers (e.g., UCSC Genome Browser)
Genome browsers are bioinformatics tools used to visualize and explore annotated genome sequences. They provide a graphical interface for navigating and analyzing genomic data, such as gene structures, regulatory elements, genetic variation, and comparative genomics. Here’s an introduction to some commonly used genome browsers:
- UCSC Genome Browser: The UCSC Genome Browser is one of the most widely used genome browsers, providing access to a wide range of genomes from various organisms. It offers a user-friendly interface for visualizing genomic features, including gene annotations, regulatory elements, and comparative genomics data. Users can navigate the genome, zoom in and out, and access detailed information about specific regions of interest.
- Ensembl Genome Browser: Ensembl is another popular genome browser that provides access to annotated genome sequences for a wide range of species. It offers similar features to the UCSC Genome Browser, including gene annotations, comparative genomics data, and tools for visualizing and analyzing genomic data.
- NCBI Genome Data Viewer: The NCBI Genome Data Viewer is a genome browser provided by the National Center for Biotechnology Information (NCBI). It allows users to visualize and explore annotated genome sequences, gene annotations, and genetic variation data from NCBI’s databases.
- JBrowse: JBrowse is a lightweight, web-based genome browser that is highly customizable and can be easily integrated into websites and applications. It offers features for visualizing and exploring genomic data, including gene annotations, sequence alignments, and genetic variation data.
- IGV (Integrative Genomics Viewer): IGV is a desktop genome browser that provides advanced features for visualizing and analyzing genomic data. It offers support for a wide range of genomic data types, including aligned sequencing reads, gene annotations, and epigenetic data.
Genome browsers are valuable tools for researchers studying genomics, genetics, and related fields, as they provide a visual representation of complex genomic data that can help in understanding the structure, function, and evolution of genomes.

Data Representation and Manipulation in Biology
Data formats used in bioinformatics (e.g., FASTA, BED, VCF)
Bioinformatics uses several standardized data formats to represent biological data, such as DNA, RNA, and protein sequences, genomic coordinates, and genetic variation. Here are some commonly used data formats in bioinformatics:
- FASTA: The FASTA format is used to represent nucleotide or amino acid sequences. It consists of a header line starting with a “>” symbol, followed by the sequence data.
Example:
shell>Sequence_1
ATCGATCGATCGATCG
- BED (Browser Extensible Data): The BED format is used to represent genomic features, such as gene annotations, transcription factor binding sites, and chromatin modifications. It consists of columns specifying the chromosome, start and end positions, and optional additional information.
Example:
yamlchr1 1000 2000 GeneA 0 +
chr1 5000 6000 GeneB 0 -
- GFF (General Feature Format): The GFF format is similar to BED and is used to represent genomic features and annotations. It includes columns for the chromosome, source, feature type, start and end positions, score, strand, and additional attributes.
Example:
yamlchr1 source gene 1000 2000 . + . gene_id=GeneA
chr1 source gene 5000 6000 . - . gene_id=GeneB
- VCF (Variant Call Format): The VCF format is used to represent genetic variation data, such as single nucleotide polymorphisms (SNPs), insertions, deletions, and structural variants. It includes columns for the chromosome, position, ID, reference allele, alternate allele(s), quality score, and additional information.
Example:
yaml#CHROM POS ID REF ALT QUAL FILTER INFO
chr1 1000 . A T 30 PASS .
chr1 5000 . G C 20 PASS .
- SAM/BAM (Sequence Alignment/Map): SAM and its binary counterpart BAM are formats used to represent aligned sequencing reads to a reference genome. SAM is a text-based format, while BAM is a binary format that is more compact and efficient for large datasets.
These are just a few examples of the many data formats used in bioinformatics. Standardized formats enable researchers to share and analyze biological data efficiently and accurately across different platforms and tools.
Introduction to data preprocessing and cleaning techniques
Data preprocessing and cleaning are essential steps in the data analysis pipeline, particularly in bioinformatics, where datasets are often large, complex, and noisy. These steps aim to prepare the data for further analysis by addressing issues such as missing values, outliers, and inconsistencies. Here’s an introduction to some common data preprocessing and cleaning techniques:
- Handling Missing Values: Missing values are common in biological datasets and can arise due to various reasons, such as experimental error or data collection issues. Common techniques for handling missing values include imputation (replacing missing values with estimated values), deletion of rows or columns with missing values, or using algorithms that can handle missing values.
- Removing Duplicates: Duplicates in a dataset can skew the analysis results. Removing duplicates ensures that each data point is unique and avoids redundancy. Duplicates can be identified based on a subset of columns or the entire row.
- Handling Outliers: Outliers are data points that significantly deviate from the rest of the dataset. Outliers can be detected using statistical methods (e.g., z-score, IQR) and then either removed or transformed (e.g., winsorization, transformation).
- Normalization and Standardization: Normalization and standardization are techniques used to scale numerical features to a standard range. Normalization scales the values to a range of [0, 1], while standardization scales the values to have a mean of 0 and a standard deviation of 1.
- Encoding Categorical Variables: Categorical variables are variables that represent categories rather than numerical values. They need to be encoded into numerical values before they can be used in machine learning algorithms. Common encoding techniques include one-hot encoding and label encoding.
- Feature Selection: Feature selection involves selecting the most relevant features (columns) from a dataset. This helps reduce the dimensionality of the dataset and improve the performance of machine learning algorithms.
- Data Transformation: Data transformation techniques, such as log transformation, square root transformation, or Box-Cox transformation, can be used to make the data distribution more Gaussian-like, which can improve the performance of some algorithms.
- Handling Skewed Data: Skewed data refers to data that is not evenly distributed. Skewed data can be transformed using techniques such as log transformation or power transformation to make the distribution more symmetric.
These are just a few examples of the many data preprocessing and cleaning techniques used in bioinformatics and data science. The choice of technique depends on the nature of the data and the specific goals of the analysis.
Overview of data visualization methods in biology
Data visualization is crucial in biology for interpreting complex datasets, identifying patterns, and communicating results effectively. Here’s an overview of some common data visualization methods used in biology:
- Bar Charts: Bar charts are used to represent categorical data, such as the frequency of different categories or groups. They are useful for comparing data across categories.
- Histograms: Histograms are used to represent the distribution of continuous data. They show the frequency of data points within each interval or bin.
- Scatter Plots: Scatter plots are used to visualize the relationship between two continuous variables. Each data point is represented as a dot, and the pattern of dots can reveal correlations or trends in the data.
- Line Graphs: Line graphs are used to show trends over time or other continuous variables. They are particularly useful for visualizing changes in data over a period of time.
- Heatmaps: Heatmaps are used to visualize large datasets by representing data values as colors. They are often used to show gene expression patterns or other large-scale biological data.
- Box Plots: Box plots are used to visualize the distribution of data and identify outliers. They show the median, quartiles, and outliers of a dataset.
- Volcano Plots: Volcano plots are used in genomics to visualize the results of differential gene expression analysis. They plot the log2 fold change on the x-axis and the -log10 of the p-value on the y-axis, with differentially expressed genes highlighted.
- Network Diagrams: Network diagrams are used to visualize complex relationships between biological entities, such as genes, proteins, or metabolites. Nodes represent entities, and edges represent relationships between them.
- Geospatial Maps: Geospatial maps are used to visualize biological data that has a spatial component, such as the distribution of species or environmental factors.
- 3D Visualization: 3D visualization techniques are used to represent biological structures, such as proteins or cells, in three-dimensional space. They are useful for understanding the spatial relationships between different components.
These are just a few examples of the many data visualization methods used in biology. The choice of visualization method depends on the nature of the data and the specific goals of the analysis.
Introduction to Computational Biology Algorithms
Pairwise sequence alignment algorithms (e.g., Needleman-Wunsch, Smith-Waterman)
Pairwise sequence alignment algorithms are used to align two sequences of nucleotides or amino acids to identify regions of similarity or homology. Two widely used algorithms for pairwise sequence alignment are the Needleman-Wunsch algorithm and the Smith-Waterman algorithm. Here’s an overview of each algorithm:
- Needleman-Wunsch Algorithm:
- Objective: The Needleman-Wunsch algorithm performs global sequence alignment, meaning it aligns the entire length of both sequences.
- Scoring: It uses a scoring matrix (e.g., BLOSUM, PAM) to assign scores to matches, mismatches, and gaps.
- Algorithm: The algorithm creates a matrix (often called the dynamic programming matrix) where each cell represents the best alignment score up to that point. It fills in the matrix by considering three possible ways to extend the alignment: extending from the cell to the left (gap in first sequence), extending from the cell above (gap in second sequence), or aligning the current characters in both sequences.
- Traceback: After filling the matrix, the algorithm traces back through the matrix from the bottom-right cell to the top-left cell to determine the optimal alignment.

Smith-Waterman Algorithm:
- Objective: The Smith-Waterman algorithm performs local sequence alignment, meaning it identifies regions of similarity within the sequences.
- Scoring: It uses a similar scoring scheme to the Needleman-Wunsch algorithm but includes an option to penalize negative scores (i.e., setting them to zero), which allows the algorithm to find local alignments.
- Algorithm: The algorithm is similar to the Needleman-Wunsch algorithm but starts the alignment from the highest-scoring cell in the matrix (i.e., the local alignment), rather than from the bottom-right corner.
- Traceback: After filling the matrix, the algorithm traces back from the highest-scoring cell to find the local alignment.
Both algorithms are based on dynamic programming and are computationally intensive, especially for long sequences. They are fundamental tools in bioinformatics for comparing biological sequences and identifying regions of conservation or functional significance.

Multiple sequence alignment algorithms (e.g., progressive alignment, iterative alignment)
Multiple sequence alignment (MSA) algorithms are used to align three or more biological sequences simultaneously to identify regions of similarity or homology. Here are two common approaches to MSA:
- Progressive Alignment:
- Algorithm: The progressive alignment algorithm constructs a guide tree that represents the evolutionary relationships between the sequences. It starts by aligning the most similar pair of sequences based on a pairwise alignment algorithm (e.g., Needleman-Wunsch or Smith-Waterman). Then, it progressively aligns additional sequences to the growing alignment, following the guide tree.
- Consistency: One challenge of progressive alignment is that errors in early alignments can propagate and lead to inaccuracies in the final alignment. Methods like profile-profile alignment can help address this issue by incorporating information from multiple alignments.
- Iterative Alignment:
- Algorithm: The iterative alignment algorithm starts with an initial alignment, which can be generated using a simple method like pairwise alignment or by using a subset of sequences. It then iteratively refines the alignment by realigning the sequences based on a more sophisticated scoring function (e.g., including position-specific scoring matrices or hidden Markov models).
- Improving Accuracy: Iterative alignment algorithms are often more computationally intensive than progressive alignment but can lead to more accurate alignments, especially for divergent sequences.
Both progressive and iterative alignment algorithms are widely used in bioinformatics for comparing biological sequences, identifying conserved regions, and inferring evolutionary relationships. The choice of algorithm depends on the specific characteristics of the sequences being aligned and the desired balance between alignment accuracy and computational efficiency.

Introduction to phylogenetic analysis algorithms
Phylogenetic analysis is the study of evolutionary relationships among groups of organisms or sequences. Phylogenetic analysis algorithms aim to reconstruct the evolutionary history, or phylogeny, of a set of sequences or taxa based on their similarities and differences. Here’s an overview of some common phylogenetic analysis algorithms:
- Distance-Based Methods:
- Neighbor-Joining (NJ): The Neighbor-Joining algorithm constructs a phylogenetic tree by iteratively joining the most similar pairs of taxa based on a distance matrix until a complete tree is formed. It is fast and computationally efficient but may not always produce the most accurate tree, especially for highly divergent sequences.
- UPGMA (Unweighted Pair Group Method with Arithmetic Mean): UPGMA is another distance-based method that constructs a tree by clustering taxa based on their pairwise distances and then iteratively joining clusters based on their average distances. It is commonly used for hierarchical clustering but is less accurate than NJ for phylogenetic analysis.
- Character-Based Methods:
- Maximum Parsimony (MP): The Maximum Parsimony algorithm reconstructs a phylogenetic tree by minimizing the number of evolutionary changes (e.g., mutations, insertions, deletions) required to explain the observed sequence data. It assumes that the most parsimonious tree (i.e., the tree with the fewest changes) is the most likely tree.
- Maximum Likelihood (ML): The Maximum Likelihood algorithm estimates the likelihood of different phylogenetic trees given the sequence data and selects the tree with the highest likelihood as the best tree. It considers the probability of observing the sequence data under different evolutionary models and tree topologies.
- Bayesian Methods:
- Bayesian Inference (BI): Bayesian Inference reconstructs a phylogenetic tree based on the posterior probability distribution of trees given the sequence data and a prior probability distribution. It uses Markov Chain Monte Carlo (MCMC) methods to sample trees from the posterior distribution and estimates the posterior probability of different tree topologies.
Phylogenetic analysis algorithms are used in various fields of biology, including evolutionary biology, microbiology, and ecology, to study the evolutionary relationships and history of organisms. Each algorithm has its strengths and limitations, and the choice of algorithm depends on the characteristics of the data and the goals of the analysis.

Case Studies and Applications
Applications of bioinformatics in various fields (e.g., medicine, agriculture, ecology)
Bioinformatics plays a crucial role in various fields, including medicine, agriculture, ecology, and beyond. Here’s an overview of some key applications of bioinformatics in these fields:
- Medicine:
- Genomic Medicine: Bioinformatics is used in genomic medicine to analyze and interpret genomic data for personalized medicine, including identifying genetic variants associated with diseases, predicting drug responses, and understanding disease mechanisms.
- Clinical Genomics: Bioinformatics tools are used in clinical settings for genetic testing, disease diagnosis, and prognosis, as well as for monitoring treatment responses and disease progression.
- Pharmacogenomics: Bioinformatics is used to study the relationship between genetic variations and drug responses, helping to develop personalized treatments and optimize drug therapies.
- Agriculture:
- Crop Improvement: Bioinformatics is used in crop improvement programs to identify genetic variations associated with desirable traits, such as yield, disease resistance, and stress tolerance, enabling the development of improved crop varieties.
- Livestock Genomics: Bioinformatics is used in livestock genomics to study the genetic basis of traits related to productivity, disease resistance, and quality of animal products, leading to the development of improved breeding strategies.
- Plant Pathogen Detection: Bioinformatics tools are used to identify and characterize plant pathogens, such as viruses, bacteria, and fungi, aiding in the development of strategies for disease management and control.
- Ecology:
- Metagenomics: Bioinformatics is used in metagenomics to study microbial communities in environmental samples, such as soil, water, and air, providing insights into microbial diversity, functions, and interactions in ecosystems.
- Environmental Genomics: Bioinformatics tools are used to study the impact of environmental factors, such as pollution and climate change, on ecosystems and biodiversity, helping to inform conservation and management strategies.
- Evolutionary Ecology: Bioinformatics is used in evolutionary ecology to study the genetic basis of adaptation and speciation, as well as to reconstruct evolutionary relationships among species.
- Other Fields:
- Biotechnology: Bioinformatics is used in biotechnology for the design of enzymes, proteins, and other biological molecules with specific functions, as well as for the optimization of metabolic pathways for biofuel production and other applications.
- Forensics: Bioinformatics tools are used in forensic genetics for DNA profiling and identification of individuals, as well as for studying genetic relationships among individuals in forensic investigations.
These are just a few examples of the wide-ranging applications of bioinformatics in various fields, highlighting its importance in advancing research, innovation, and applications in biology and related disciplines.
Case studies highlighting the integration of biology and computer science in solving biological problems
Here are a few case studies that illustrate the integration of biology and computer science in solving biological problems:
- Genome Assembly and Annotation:
- Problem: Sequencing the human genome generates vast amounts of raw sequencing data that need to be assembled into a complete genome and annotated to identify genes and regulatory elements.
- Solution: Bioinformatics algorithms and software, such as BLAST, Velvet, and AUGUSTUS, are used to assemble and annotate genomes by aligning sequencing reads, predicting gene structures, and identifying functional elements.
- Phylogenetic Analysis:
- Problem: Reconstructing the evolutionary relationships among species based on genetic data requires comparing and analyzing large datasets of sequences.
- Solution: Phylogenetic analysis tools, such as PHYLIP, MEGA, and MrBayes, use computational algorithms to infer phylogenetic trees from sequence data, helping to understand evolutionary history and biodiversity.
- Drug Discovery and Design:
- Problem: Identifying potential drug targets and designing new drugs requires analyzing the structure and function of biological molecules, such as proteins and nucleic acids.
- Solution: Computational tools, such as molecular docking software (e.g., AutoDock, GOLD) and molecular dynamics simulations, are used to predict the interactions between drugs and target molecules, aiding in drug discovery and design.
- Metagenomics and Microbiome Analysis:
- Problem: Studying complex microbial communities in environmental and clinical samples requires analyzing metagenomic data to identify and characterize individual microbial species.
- Solution: Bioinformatics pipelines, such as QIIME, MetaPhlAn, and MG-RAST, are used to process and analyze metagenomic data, providing insights into microbial diversity, functions, and interactions.
- Protein Structure Prediction:
- Problem: Predicting the three-dimensional structure of proteins from their amino acid sequences is essential for understanding their functions and interactions.
- Solution: Computational methods, such as homology modeling, ab initio modeling, and machine learning-based approaches, are used to predict protein structures, aiding in drug discovery, protein engineering, and functional annotation.
These case studies highlight how the integration of biology and computer science, through the use of bioinformatics tools and algorithms, can address complex biological problems and advance our understanding of biological systems.
Ethical and Legal Considerations in Bioinformatics
Ethical issues related to bioinformatics research (e.g., data privacy, informed consent)
Ethical issues in bioinformatics research arise from the use of large-scale biological data, such as genomic and clinical data, and the potential implications for individuals, communities, and society. Some key ethical issues in bioinformatics research include:
- Data Privacy and Security: Bioinformatics research often involves the use of sensitive personal data, such as genomic information and health records. Ensuring the privacy and security of this data is crucial to protect individuals from unauthorized access and misuse.
- Informed Consent: Obtaining informed consent from individuals whose data is used in research is essential to ensure that they understand the nature of the research, its potential risks and benefits, and how their data will be used and shared.
- Data Sharing and Access: Balancing the need for data sharing to advance scientific knowledge with the protection of individual privacy and confidentiality is a complex ethical issue. Researchers must ensure that data sharing practices are transparent, and data are de-identified or anonymized to protect privacy.
- Equity and Access: Ensuring equitable access to bioinformatics tools, data, and research findings is essential to avoid exacerbating existing disparities in healthcare and research opportunities.
- Ethical Use of AI and Machine Learning: As bioinformatics increasingly relies on artificial intelligence (AI) and machine learning (ML) algorithms, ensuring that these technologies are used ethically and do not perpetuate biases is critical.
- Responsible Conduct of Research: Adhering to ethical standards and guidelines for research conduct, such as the Declaration of Helsinki and the Belmont Report, is essential to maintain the integrity and trustworthiness of bioinformatics research.
- Dual-Use Research: Bioinformatics research can have both beneficial and potentially harmful applications. Researchers must consider the potential dual-use implications of their work and take steps to minimize the risks of misuse.
- Community Engagement and Public Communication: Engaging with communities affected by bioinformatics research and communicating research findings responsibly to the public are essential for building trust and ensuring that research is conducted in the public interest.
Addressing these ethical issues requires collaboration among researchers, policymakers, and the public to develop and implement ethical guidelines and best practices that protect individuals’ rights and promote the responsible conduct of bioinformatics research.
Legal considerations (e.g., intellectual property rights, data sharing policies)
Legal considerations in bioinformatics research encompass a range of issues related to intellectual property rights, data sharing policies, regulatory compliance, and ethical standards. Some key legal considerations in bioinformatics research include:
- Intellectual Property Rights (IPR): Bioinformatics tools, algorithms, and databases may be eligible for intellectual property protection, such as patents, copyrights, or trade secrets. Researchers should be aware of IPR laws and regulations to protect their innovations and respect the rights of others.
- Data Sharing Policies: Many funding agencies, journals, and institutions require researchers to share data, software, and other research outputs to promote transparency and reproducibility. Researchers should comply with data sharing policies and ensure that data are shared responsibly and ethically.
- Privacy and Data Protection: Researchers working with sensitive personal data, such as genomic information or health records, must comply with data protection laws and regulations to ensure the privacy and security of individuals’ data.
- Regulatory Compliance: Bioinformatics research involving human subjects, animals, or hazardous materials may be subject to regulatory requirements, such as ethics approvals, animal care and use protocols, and biosafety regulations. Researchers should comply with relevant regulations to ensure the safety and welfare of research participants and the public.
- Ethical Standards: Bioinformatics researchers should adhere to ethical standards and guidelines, such as those outlined in the Declaration of Helsinki and the Belmont Report, to ensure the welfare of research participants, the integrity of research findings, and the responsible conduct of research.
- Commercialization and Technology Transfer: Researchers who develop bioinformatics tools or technologies with commercial potential should be aware of technology transfer and commercialization processes, including licensing agreements and intellectual property management.
- International Collaboration: Bioinformatics research often involves collaboration across borders, raising legal considerations related to data transfer, intellectual property rights, and regulatory compliance in different jurisdictions.
- Open Science and Open Access: Embracing open science principles, such as open access publishing and open data sharing, can enhance the visibility and impact of bioinformatics research but may also raise legal considerations related to licensing and attribution.
Navigating these legal considerations requires a comprehensive understanding of relevant laws, regulations, and policies, as well as effective communication and collaboration with legal advisors, institutional officials, and other stakeholders.
Future Directions in Bioinformatics
Emerging trends and technologies in bioinformatics
Emerging trends and technologies in bioinformatics are shaping the future of biological research and healthcare. Some key trends include:
- Single-cell Omics: Advances in single-cell sequencing technologies are enabling researchers to study the molecular profiles of individual cells, providing insights into cellular heterogeneity, developmental processes, and disease mechanisms.
- Spatial Omics: Spatial omics technologies, such as spatial transcriptomics and spatial proteomics, allow researchers to study the spatial organization of biomolecules within tissues, providing spatial context to molecular data.
- Long-read Sequencing: Long-read sequencing technologies, such as Oxford Nanopore and PacBio, are revolutionizing genome sequencing by enabling the assembly of complex genomes, detection of structural variants, and characterization of repetitive regions.
- AI and Machine Learning: AI and machine learning algorithms are increasingly used in bioinformatics for data analysis, pattern recognition, and predictive modeling, enabling researchers to extract meaningful insights from large and complex biological datasets.
- CRISPR-based Technologies: CRISPR-Cas9 and other CRISPR-based technologies are revolutionizing genetic engineering, enabling precise genome editing and the study of gene function and regulation.
- Multi-omics Integration: Integrating multiple omics datasets, such as genomics, transcriptomics, proteomics, and metabolomics, is enabling researchers to gain a more comprehensive understanding of biological systems and diseases.
- Personalized Medicine: Bioinformatics is playing a key role in personalized medicine by enabling the analysis of individual genomes, identification of genetic markers for disease risk, and development of targeted therapies.
- Data Sharing and Collaboration: There is a growing emphasis on data sharing and collaboration in bioinformatics, facilitated by initiatives such as the Global Alliance for Genomics and Health (GA4GH) and the FAIR (Findable, Accessible, Interoperable, and Reusable) data principles.
- Ethical and Legal Considerations: As bioinformatics technologies advance, there is an increasing focus on addressing ethical and legal issues related to data privacy, informed consent, and responsible use of technology.
These trends are driving innovation in bioinformatics and have the potential to revolutionize our understanding of biology, improve healthcare outcomes, and address global challenges in health and the environment.
Career paths and opportunities in bioinformatics
Bioinformatics offers a wide range of career paths and opportunities for individuals interested in combining biology, computer science, and data analysis. Some common career paths in bioinformatics include:
- Bioinformatics Scientist/Analyst: Bioinformatics scientists and analysts work in research institutions, biotech companies, pharmaceutical companies, and government agencies. They analyze biological data, develop algorithms and software tools, and interpret results to answer biological questions.
- Computational Biologist: Computational biologists use computational tools and models to study biological systems, analyze biological data, and develop new algorithms and methods for biological research.
- Data Scientist: Data scientists in bioinformatics work with large and complex biological datasets, using statistical analysis, machine learning, and other data analysis techniques to extract meaningful insights and patterns.
- Biostatistician: Biostatisticians apply statistical methods to analyze biological data, design experiments, and interpret results in areas such as genomics, epidemiology, and clinical trials.
- Software Engineer/Developer: Software engineers and developers in bioinformatics design, develop, and maintain software tools, databases, and applications for analyzing biological data and conducting research.
- Research Scientist: Research scientists in bioinformatics lead research projects, develop new algorithms and methods, publish scientific papers, and collaborate with other researchers to advance knowledge in the field.
- Clinical Bioinformatician: Clinical bioinformaticians work in healthcare settings, applying bioinformatics tools and methods to analyze patient data, diagnose diseases, predict treatment outcomes, and personalize medicine.
- Academic/Research Faculty: Many bioinformatics professionals pursue academic careers, becoming professors or research faculty at universities, where they conduct research, teach courses, and mentor students.
- Entrepreneur/Startup Founder: Some bioinformatics professionals start their own companies or join startups to develop innovative bioinformatics tools, technologies, and services for the biotech and healthcare industries.
- Consultant: Bioinformatics consultants provide expertise and guidance to organizations on bioinformatics-related projects, helping them solve complex biological problems and optimize research processes.
Overall, bioinformatics offers a diverse range of career paths and opportunities for individuals with a passion for biology, computer science, and data analysis. The field is dynamic and rapidly evolving, with new technologies and applications continuing to expand its scope and impact.


